Myeloid cell-specific deletion of epidermal growth factor receptor aggravates acute cardiac injury
- PMID: 37728308
- PMCID: PMC10758753
- DOI: 10.1042/CS20230804
Myeloid cell-specific deletion of epidermal growth factor receptor aggravates acute cardiac injury
Abstract
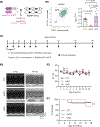
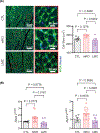
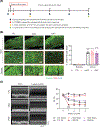
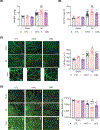
Myeloid cells, including macrophages, play important roles as first responders to cardiac injury and stress. Epidermal growth factor receptor (EGFR) has been identified as a mediator of macrophage responsiveness to select diseases, though its impact on cardiac function or remodeling following acute ischemic injury is unknown. We aimed to define the role of myeloid cell-specific EGFR in the regulation of cardiac function and remodeling following acute myocardial infarction (MI)-induced injury. Floxed EGFR mice were bred with homozygous LysM-Cre (LMC) transgenic mice to yield myeloid-specific EGFR knockout (mKO) mice. Via echocardiography, immunohistochemistry, RNA sequencing and flow cytometry, the impact of myeloid cell-specific EGFR deletion on cardiac structure and function was assessed at baseline and following injury. Compared with LMC controls, myeloid cell-specific EGFR deletion led to an increase in cardiomyocyte hypertrophy at baseline. Bulk RNASeq analysis of isolated cardiac Cd11b+ myeloid cells revealed substantial changes in mKO cell transcripts at baseline, particularly in relation to predicted decreases in neovascularization. In response to myocardial infarction, mKO mice experienced a hastened decline in cardiac function with isolated cardiac Cd11b+ myeloid cells expressing decreased levels of the pro-reparative mediators Vegfa and Il10, which coincided with enhanced cardiac hypertrophy and decreased capillary density. Overall, loss of EGFR qualitatively alters cardiac resident macrophages that promotes a low level of basal stress and a more rapid decrease in cardiac function along with worsened repair following acute ischemic injury.
Keywords: epidermal growth factor receptor; macrophages; myeloid cell; myocardial remodeling.
© 2023 The Author(s). Published by Portland Press Limited on behalf of the Biochemical Society.
Figures

Similar articles
-
Amphiregulin enhances cardiac fibrosis and aggravates cardiac dysfunction in mice with experimental myocardial infarction partly through activating EGFR-dependent pathway.Basic Res Cardiol. 2018 Jan 18;113(2):12. doi: 10.1007/s00395-018-0669-y. Basic Res Cardiol. 2018. Retraction in: Basic Res Cardiol. 2022 Nov 7;117(1):54. doi: 10.1007/s00395-022-00963-2. PMID: 29349588 Retracted.
-
Myeloid-Epithelial-Reproductive Receptor Tyrosine Kinase and Milk Fat Globule Epidermal Growth Factor 8 Coordinately Improve Remodeling After Myocardial Infarction via Local Delivery of Vascular Endothelial Growth Factor.Circulation. 2016 Mar 1;133(9):826-39. doi: 10.1161/CIRCULATIONAHA.115.020857. Epub 2016 Jan 27. Circulation. 2016. PMID: 26819373 Free PMC article.
-
Activation of the epidermal growth factor receptor in macrophages regulates cytokine production and experimental colitis.J Immunol. 2014 Feb 1;192(3):1013-23. doi: 10.4049/jimmunol.1300133. Epub 2014 Jan 3. J Immunol. 2014. PMID: 24391216 Free PMC article.
-
Iron Regulator Hepcidin Impairs Macrophage-Dependent Cardiac Repair After Injury.Circulation. 2019 Mar 19;139(12):1530-1547. doi: 10.1161/CIRCULATIONAHA.118.034545. Circulation. 2019. PMID: 30586758
-
Cardiac macrophages and emerging roles for their metabolism after myocardial infarction.J Clin Invest. 2023 Sep 15;133(18):e171953. doi: 10.1172/JCI171953. J Clin Invest. 2023. PMID: 37712418 Free PMC article. Review.
Cited by
-
Loss of cardiomyocyte-specific adhesion G-protein-coupled receptor G1 (ADGRG1/GPR56) promotes pressure overload-induced heart failure.Biosci Rep. 2024 Sep 25;44(9):BSR20240826. doi: 10.1042/BSR20240826. Biosci Rep. 2024. PMID: 39264336 Free PMC article.
-
The immune system in cardiovascular diseases: from basic mechanisms to therapeutic implications.Signal Transduct Target Ther. 2025 May 23;10(1):166. doi: 10.1038/s41392-025-02220-z. Signal Transduct Target Ther. 2025. PMID: 40404619 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

