VirB, a key transcriptional regulator of Shigella virulence, requires a CTP ligand for its regulatory activities
- PMID: 37728345
- PMCID: PMC10653881
- DOI: 10.1128/mbio.01519-23
VirB, a key transcriptional regulator of Shigella virulence, requires a CTP ligand for its regulatory activities
Abstract
Shigella species cause bacillary dysentery, the second leading cause of diarrheal deaths worldwide. There is a pressing need to identify novel molecular drug targets. Shigella virulence phenotypes are controlled by the transcriptional regulator, VirB. We show that VirB belongs to a fast-evolving, plasmid-borne clade of the ParB superfamily, which has diverged from versions with a distinct cellular role-DNA partitioning. We report that, like classic members of the ParB family, VirB binds a highly unusual ligand, CTP. Mutants predicted to be defective in CTP binding are compromised in a variety of virulence attributes controlled by VirB, likely because these mutants cannot engage DNA. This study (i) reveals that VirB binds CTP, (ii) provides a link between VirB-CTP interactions and Shigella virulence phenotypes, (iii) provides new insight into VirB-CTP-DNA interactions, and (iv) broadens our understanding of the ParB superfamily, a group of bacterial proteins that play critical roles in many bacteria.
Keywords: Congo red phenotype; GFP fusions; ParB/Spo0J; bacterial gene regulation; focus formation; large plasmids; plasmid partitioning; transcriptional silencing/anti-silencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

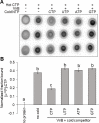

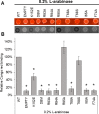

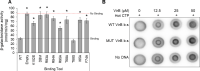
Update of
-
VirB, a key transcriptional regulator of Shigella virulence, requires a CTP ligand for its regulatory activities.bioRxiv [Preprint]. 2023 May 18:2023.05.16.541010. doi: 10.1101/2023.05.16.541010. bioRxiv. 2023. Update in: mBio. 2023 Oct 31;14(5):e0151923. doi: 10.1128/mbio.01519-23. PMID: 37293012 Free PMC article. Updated. Preprint.
References
-
- Niyogi SK. 2005. Shigellosis. J Microbiol 43:133–143. - PubMed
-
- Weatherspoon-Griffin N, Picker MA, Wing HJ. 2016. The genetic organization and transcriptional regulation of Shigella virulence genes, p 65–107. In Picking WD, Picking WL (ed), Shigella: Molecular and cellular biology. Caister Academic Press, UK. doi:10.21775/9781910190197 - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
