Characterization of a family I inorganic pyrophosphatase from Legionella pneumophila Philadelphia 1
- PMID: 37728609
- PMCID: PMC10565794
- DOI: 10.1107/S2053230X23008002
Characterization of a family I inorganic pyrophosphatase from Legionella pneumophila Philadelphia 1
Abstract
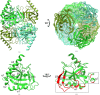
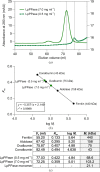
Inorganic pyrophosphate (PPi) is generated as an intermediate or byproduct of many fundamental metabolic pathways, including DNA/RNA synthesis. The intracellular concentration of PPi must be regulated as buildup can inhibit many critical cellular processes. Inorganic pyrophosphatases (PPases) hydrolyze PPi into two orthophosphates (Pi), preventing the toxic accumulation of the PPi byproduct in cells and making Pi available for use in biosynthetic pathways. Here, the crystal structure of a family I inorganic pyrophosphatase from Legionella pneumophila is reported at 2.0 Å resolution. L. pneumophila PPase (LpPPase) adopts a homohexameric assembly and shares the oligonucleotide/oligosaccharide-binding (OB) β-barrel core fold common to many other bacterial family I PPases. LpPPase demonstrated hydrolytic activity against a general substrate, with Mg2+ being the preferred metal cofactor for catalysis. Legionnaires' disease is a severe respiratory infection caused primarily by L. pneumophila, and thus increased characterization of the L. pneumophila proteome is of interest.
Keywords: Legionella pneumophila; Legionnaires' disease; SSGCID; Seattle Structural Genomics Center for Infectious Disease; inorganic pyrophosphatases; structural genomics.
Figures


Similar articles
-
Induced expression of the Legionella pneumophila gene encoding a 20-kilodalton protein during intracellular infection.Infect Immun. 1998 Jan;66(1):203-12. doi: 10.1128/IAI.66.1.203-212.1998. Infect Immun. 1998. PMID: 9423859 Free PMC article.
-
A Supervised Statistical Learning Approach for Accurate Legionella pneumophila Source Attribution during Outbreaks.Appl Environ Microbiol. 2017 Oct 17;83(21):e01482-17. doi: 10.1128/AEM.01482-17. Print 2017 Nov 1. Appl Environ Microbiol. 2017. PMID: 28821546 Free PMC article.
-
Characterization of Legionella pneumophila isolated from environmental water and ashiyu foot spa.Biomed Res Int. 2013;2013:514395. doi: 10.1155/2013/514395. Epub 2013 Jul 11. Biomed Res Int. 2013. PMID: 23956987 Free PMC article.
-
Legionella pneumophila type IV effectors hijack the transcription and translation machinery of the host cell.Trends Cell Biol. 2014 Dec;24(12):771-8. doi: 10.1016/j.tcb.2014.06.002. Epub 2014 Jul 8. Trends Cell Biol. 2014. PMID: 25012125 Review.
-
Molecular pathogenesis of infections caused by Legionella pneumophila.Clin Microbiol Rev. 2010 Apr;23(2):274-98. doi: 10.1128/CMR.00052-09. Clin Microbiol Rev. 2010. PMID: 20375353 Free PMC article. Review.
Cited by
-
Using structural genomics depositions in undergraduate teaching of protein crystallography: everybody wins.Acta Crystallogr F Struct Biol Commun. 2023 Oct 1;79(Pt 10):274-275. doi: 10.1107/S2053230X2300883X. Epub 2023 Oct 1. Acta Crystallogr F Struct Biol Commun. 2023. PMID: 37815477 Free PMC article.
-
Structures of Legionella pneumophila serogroup 1 peptide deformylase bound to nickel(II) and actinonin.Acta Crystallogr F Struct Biol Commun. 2025 Apr 1;81(Pt 4):163-170. doi: 10.1107/S2053230X25001876. Epub 2025 Mar 17. Acta Crystallogr F Struct Biol Commun. 2025. PMID: 40091854
-
Structural and functional characterization of FabG4 from Mycolicibacterium smegmatis.Acta Crystallogr F Struct Biol Commun. 2024 Apr 1;80(Pt 4):82-91. doi: 10.1107/S2053230X2400356X. Epub 2024 Apr 24. Acta Crystallogr F Struct Biol Commun. 2024. PMID: 38656226 Free PMC article.
-
Structural characterization of dUTPase from Legionella pneumophila.Acta Crystallogr F Struct Biol Commun. 2025 Apr 1;81(Pt 4):155-162. doi: 10.1107/S2053230X25001815. Epub 2025 Mar 17. Acta Crystallogr F Struct Biol Commun. 2025. PMID: 40091853
-
Carbohydrate Deacetylase Unique to Gut Microbe Bacteroides Reveals Atypical Structure.Biochemistry. 2025 Jan 7;64(1):180-191. doi: 10.1021/acs.biochem.4c00519. Epub 2024 Dec 11. Biochemistry. 2025. PMID: 39663570 Free PMC article.
References
-
- Agirre, J., Atanasova, M., Bagdonas, H., Ballard, C. B., Baslé, A., Beilsten-Edmands, J., Borges, R. J., Brown, D. G., Burgos-Mármol, J. J., Berrisford, J. M., Bond, P. S., Caballero, I., Catapano, L., Chojnowski, G., Cook, A. G., Cowtan, K. D., Croll, T. I., Debreczeni, J. É., Devenish, N. E., Dodson, E. J., Drevon, T. R., Emsley, P., Evans, G., Evans, P. R., Fando, M., Foadi, J., Fuentes-Montero, L., Garman, E. F., Gerstel, M., Gildea, R. J., Hatti, K., Hekkelman, M. L., Heuser, P., Hoh, S. W., Hough, M. A., Jenkins, H. T., Jiménez, E., Joosten, R. P., Keegan, R. M., Keep, N., Krissinel, E. B., Kolenko, P., Kovalevskiy, O., Lamzin, V. S., Lawson, D. M., Lebedev, A. A., Leslie, A. G. W., Lohkamp, B., Long, F., Malý, M., McCoy, A. J., McNicholas, S. J., Medina, A., Millán, C., Murray, J. W., Murshudov, G. N., Nicholls, R. A., Noble, M. E. M., Oeffner, R., Pannu, N. S., Parkhurst, J. M., Pearce, N., Pereira, J., Perrakis, A., Powell, H. R., Read, R. J., Rigden, D. J., Rochira, W., Sammito, M., Sánchez Rodríguez, F., Sheldrick, G. M., Shelley, K. L., Simkovic, F., Simpkin, A. J., Skubak, P., Sobolev, E., Steiner, R. A., Stevenson, K., Tews, I., Thomas, J. M. H., Thorn, A., Valls, J. T., Uski, V., Usón, I., Vagin, A., Velankar, S., Vollmar, M., Walden, H., Waterman, D., Wilson, K. S., Winn, M. D., Winter, G., Wojdyr, M. & Yamashita, K. (2023). Acta Cryst. D79, 449–461.
-
- Baykov, A. A., Dudarenkov, V. Y., Käpylä, J., Salminen, T., Hyytiä, T., Kasho, V. N., Husgafvel, S., Cooperman, B. S., Goldman, A. & Lahti, R. (1995). J. Biol. Chem. 270, 30804–30812. - PubMed
-
- Centers for Disease Control and Prevention (2022). Legionnaires’ Disease and Pontiac Fever. https://www.cdc.gov/legionella/index.html.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

