UHRF1 promotes spindle assembly and chromosome congression by catalyzing EG5 polyubiquitination
- PMID: 37728657
- PMCID: PMC10510743
- DOI: 10.1083/jcb.202210093
UHRF1 promotes spindle assembly and chromosome congression by catalyzing EG5 polyubiquitination
Abstract
UHRF1 is an epigenetic coordinator bridging DNA methylation and histone modifications. Additionally, UHRF1 regulates DNA replication and cell cycle, and its deletion induces G1/S or G2/M cell cycle arrest. The roles of UHRF1 in the regulation of G2/M transition remain poorly understood. UHRF1 depletion caused chromosome misalignment, thereby inducing cell cycle arrest at mitotic metaphase, and these cells exhibited the defects of spindle geometry, prominently manifested as shorter spindles. Mechanistically, UHRF1 protein directly interacts with EG5, a kinesin motor protein, during mitosis. Furthermore, UHRF1 induced EG5 polyubiquitination at the site of K1034 and further promoted the interaction of EG5 with spindle assembly factor TPX2, thereby ensuring accurate EG5 distribution to the spindles during metaphase. Our study clarifies a novel UHRF1 function as a nuclear protein catalyzing EG5 polyubiquitination for proper spindle architecture and faithful genomic transmission, which is independent of its roles in epigenetic regulation and DNA damage repair inside the nucleus. These findings revealed a previously unknown mechanism of UHRF1 in controlling mitotic spindle architecture and chromosome behavior and provided mechanistic evidence for UHRF1 deletion-mediated G2/M arrest.
© 2023 Qi et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures








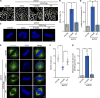
Similar articles
-
PTEN regulates EG5 to control spindle architecture and chromosome congression during mitosis.Nat Commun. 2016 Aug 5;7:12355. doi: 10.1038/ncomms12355. Nat Commun. 2016. PMID: 27492783 Free PMC article.
-
Eg5 UFMylation promotes spindle organization during mitosis.Cell Death Dis. 2024 Jul 31;15(7):544. doi: 10.1038/s41419-024-06934-w. Cell Death Dis. 2024. PMID: 39085203 Free PMC article.
-
NAT10 regulates mitotic cell fate by acetylating Eg5 to control bipolar spindle assembly and chromosome segregation.Cell Death Differ. 2022 Apr;29(4):846-860. doi: 10.1038/s41418-021-00899-5. Epub 2022 Feb 24. Cell Death Differ. 2022. PMID: 35210604 Free PMC article.
-
Non-canonical functions of the mitotic kinesin Eg5.Thorac Cancer. 2018 Aug;9(8):904-910. doi: 10.1111/1759-7714.12792. Epub 2018 Jun 21. Thorac Cancer. 2018. PMID: 29927078 Free PMC article. Review.
-
G2 and spindle assembly checkpoint adaptation, and tetraploidy arrest: implications for intrinsic and chemically induced genomic instability.Mutat Res. 2003 Nov 27;532(1-2):245-53. doi: 10.1016/j.mrfmmm.2003.08.020. Mutat Res. 2003. PMID: 14643440 Review.
Cited by
-
SKP2 inhibition activates tumor cell-intrinsic immunity by inducing DNA replication stress and genomic instability.Br J Cancer. 2025 Jan;132(1):81-92. doi: 10.1038/s41416-024-02909-y. Epub 2024 Nov 24. Br J Cancer. 2025. PMID: 39582087
-
Integrated bioinformatics combined with machine learning to analyze shared biomarkers and pathways in psoriasis and cervical squamous cell carcinoma.Front Immunol. 2024 May 28;15:1351908. doi: 10.3389/fimmu.2024.1351908. eCollection 2024. Front Immunol. 2024. PMID: 38863714 Free PMC article.
-
High interstitial fluid pressure enhances USP1-dependent KIF11 protein stability to promote hepatocellular carcinoma progression.J Transl Med. 2025 Jan 14;23(1):66. doi: 10.1186/s12967-025-06124-y. J Transl Med. 2025. PMID: 39810156 Free PMC article.
-
Roles of post-translational modifications of UHRF1 in cancer.Epigenetics Chromatin. 2024 May 9;17(1):15. doi: 10.1186/s13072-024-00540-y. Epigenetics Chromatin. 2024. PMID: 38725075 Free PMC article. Review.
-
Histone Deacetylases in Retinoblastoma.Int J Mol Sci. 2024 Jun 24;25(13):6910. doi: 10.3390/ijms25136910. Int J Mol Sci. 2024. PMID: 39000021 Free PMC article. Review.
References
-
- Arima, Y., Hirota T., Bronner C., Mousli M., Fujiwara T., Niwa S., Ishikawa H., and Saya H.. 2004. Down-regulation of nuclear protein ICBP90 by p53/p21Cip1/WAF1-dependent DNA-damage checkpoint signals contributes to cell cycle arrest at G1/S transition. Genes Cells. 9:131–142. 10.1111/j.1356-9597.2004.00710.x - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

