Characterization of Proteoform Post-Translational Modifications by Top-Down and Bottom-Up Mass Spectrometry in Conjunction with Annotations
- PMID: 37728997
- PMCID: PMC10563160
- DOI: 10.1021/acs.jproteome.3c00207
Characterization of Proteoform Post-Translational Modifications by Top-Down and Bottom-Up Mass Spectrometry in Conjunction with Annotations
Abstract
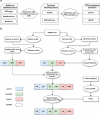
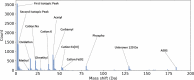
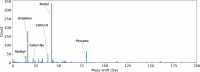
Many proteoforms can be produced from a gene due to genetic mutations, alternative splicing, post-translational modifications (PTMs), and other variations. PTMs in proteoforms play critical roles in cell signaling, protein degradation, and other biological processes. Mass spectrometry (MS) is the primary technique for investigating PTMs in proteoforms, and two alternative MS approaches, top-down and bottom-up, have complementary strengths. The combination of the two approaches has the potential to increase the sensitivity and accuracy in PTM identification and characterization. In addition, protein and PTM knowledge bases, such as UniProt, provide valuable information for PTM characterization and verification. Here, we present a software pipeline PTM-TBA (PTM characterization by Top-down and Bottom-up MS and Annotations) for identifying and localizing PTMs in proteoforms by integrating top-down and bottom-up MS as well as PTM annotations. We assessed PTM-TBA using a technical triplicate of bottom-up and top-down MS data of SW480 cells. On average, database search of the top-down MS data identified 2000 mass shifts, 814.5 (40.7%) of which were matched to 11 common PTMs and 423 of which were localized. Of the mass shifts identified by top-down MS, PTM-TBA verified 435 mass shifts using the bottom-up MS data and UniProt annotations.
Keywords: bottom-up mass spectrometry; post-translational modification; top-down mass spectrometry.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

Update of
-
Characterization of proteoform post-translational modifications by top-down and bottom-up mass spectrometry in conjunction with UniProt annotations.bioRxiv [Preprint]. 2023 Apr 6:2023.04.04.535618. doi: 10.1101/2023.04.04.535618. bioRxiv. 2023. Update in: J Proteome Res. 2023 Oct 6;22(10):3178-3189. doi: 10.1021/acs.jproteome.3c00207. PMID: 37066296 Free PMC article. Updated. Preprint.
Similar articles
-
Characterization of proteoform post-translational modifications by top-down and bottom-up mass spectrometry in conjunction with UniProt annotations.bioRxiv [Preprint]. 2023 Apr 6:2023.04.04.535618. doi: 10.1101/2023.04.04.535618. bioRxiv. 2023. Update in: J Proteome Res. 2023 Oct 6;22(10):3178-3189. doi: 10.1021/acs.jproteome.3c00207. PMID: 37066296 Free PMC article. Updated. Preprint.
-
Improving Proteoform Identifications in Complex Systems Through Integration of Bottom-Up and Top-Down Data.J Proteome Res. 2020 Aug 7;19(8):3510-3517. doi: 10.1021/acs.jproteome.0c00332. Epub 2020 Jul 10. J Proteome Res. 2020. PMID: 32584579 Free PMC article.
-
Identification of ultramodified proteins using top-down tandem mass spectra.J Proteome Res. 2013 Dec 6;12(12):5830-8. doi: 10.1021/pr400849y. Epub 2013 Nov 15. J Proteome Res. 2013. PMID: 24188097 Free PMC article.
-
Top-Down Proteomics and the Challenges of True Proteoform Characterization.J Proteome Res. 2023 Dec 1;22(12):3663-3675. doi: 10.1021/acs.jproteome.3c00416. Epub 2023 Nov 8. J Proteome Res. 2023. PMID: 37937372 Free PMC article. Review.
-
The top-down, middle-down, and bottom-up mass spectrometry approaches for characterization of histone variants and their post-translational modifications.Proteomics. 2014 Mar;14(4-5):489-97. doi: 10.1002/pmic.201300256. Epub 2013 Dec 16. Proteomics. 2014. PMID: 24339419 Review.
Cited by
-
Mass Spectrometry-Based Top-Down Proteomics in Nanomedicine: Proteoform-Specific Measurement of Protein Corona.ACS Nano. 2024 Sep 14;18(38):26024-36. doi: 10.1021/acsnano.4c04675. Online ahead of print. ACS Nano. 2024. PMID: 39276099 Free PMC article.
-
Time-lapse tryptic digestion: a proteomic approach to improve sequence coverage of extracellular matrix proteins.bioRxiv [Preprint]. 2025 Mar 28:2025.03.26.645502. doi: 10.1101/2025.03.26.645502. bioRxiv. 2025. PMID: 40196545 Free PMC article. Preprint.
-
The role of protein corona in advancing plasma proteomics.Proteomics. 2025 Jan;25(1-2):e2400028. doi: 10.1002/pmic.202400028. Epub 2024 Sep 2. Proteomics. 2025. PMID: 39221533 Free PMC article. Review.
-
Proteomic insights into the extracellular matrix: a focus on proteoforms and their implications in health and disease.Expert Rev Proteomics. 2024 Nov;21(11):463-481. doi: 10.1080/14789450.2024.2427136. Epub 2024 Nov 15. Expert Rev Proteomics. 2024. PMID: 39512072 Review.
-
Mass spectrometry-intensive top-down proteomics: an update on technology advancements and biomedical applications.Anal Methods. 2024 Jul 18;16(28):4664-4682. doi: 10.1039/d4ay00651h. Anal Methods. 2024. PMID: 38973469 Free PMC article. Review.
References
-
- Rikova K.; Guo A.; Zeng Q.; Possemato A.; Yu J.; Haack H.; Nardone J.; Lee K.; Reeves C.; Li Y.; Hu Y.; Tan Z.; Stokes M.; Sullivan L.; Mitchell J.; Wetzel R.; MacNeill J.; Ren J. M.; Yuan J.; Bakalarski C. E.; Villen J.; Kornhauser J. M.; Smith B.; Li D.; Zhou X.; Gygi S. P.; Gu T. L.; Polakiewicz R. D.; Rush J.; Comb M. J. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 2007, 131 (6), 1190–1203. 10.1016/j.cell.2007.11.025. - DOI - PubMed
-
- Wang Y.; Zhang J.; Li B.; He Q. Y. Advances of proteomics in novel PTM discovery: applications in cancer therapy. Small Methods 2019, 3 (5), 1900041.10.1002/smtd.201900041. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

