Structural transitions upon guide RNA binding and their importance in Cas12g-mediated RNA cleavage
- PMID: 37729124
- PMCID: PMC10511118
- DOI: 10.1371/journal.pgen.1010930
Structural transitions upon guide RNA binding and their importance in Cas12g-mediated RNA cleavage
Abstract
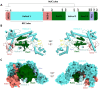
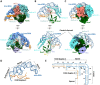
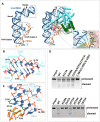
Cas12g is an endonuclease belonging to the type V RNA-guided CRISPR-Cas family. It is known for its ability to cleave RNA substrates using a conserved endonuclease active site located in the RuvC domain. In this study, we determined the crystal structure of apo-Cas12g, the cryo-EM structure of the Cas12g-sgRNA binary complex and investigated conformational changes that occur during the transition from the apo state to the Cas12g-sgRNA binary complex. The conserved zinc finger motifs in Cas12g undergo an ordered-to-disordered transition from the apo to the sgRNA-bound state and their mutations negatively impact on target RNA cleavage. Moreover, we identified a lid motif in the RuvC domain that undergoes transformation from a helix to loop to regulate the access to the RuvC active site and subsequent cleavage of the RNA substrate. Overall, our study provides valuable insights into the mechanisms by which Cas12g recognizes sgRNA and the conformational changes it undergoes from sgRNA binding to the activation of the RNase active site, thereby laying a foundation for the potential repurposing of Cas12g as a tool for RNA-editing.
Copyright: © 2023 Liu et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
A New RNA-Dependent Cas12g Nuclease.Int J Mol Sci. 2023 Dec 4;24(23):17105. doi: 10.3390/ijms242317105. Int J Mol Sci. 2023. PMID: 38069429 Free PMC article.
-
Cryo-EM structure of the RNA-guided ribonuclease Cas12g.Nat Chem Biol. 2021 Apr;17(4):387-393. doi: 10.1038/s41589-020-00721-2. Epub 2021 Jan 25. Nat Chem Biol. 2021. PMID: 33495647 Free PMC article.
-
Full-Length Model of SaCas9-sgRNA-DNA Complex in Cleavage State.Int J Mol Sci. 2023 Jan 7;24(2):1204. doi: 10.3390/ijms24021204. Int J Mol Sci. 2023. PMID: 36674715 Free PMC article.
-
Class 2 CRISPR-Cas RNA-guided endonucleases: Swiss Army knives of genome editing.Nat Struct Mol Biol. 2017 Nov;24(11):882-892. doi: 10.1038/nsmb.3486. Epub 2017 Oct 16. Nat Struct Mol Biol. 2017. PMID: 29035385 Review.
-
Computational Tools and Resources for CRISPR/Cas Genome Editing.Genomics Proteomics Bioinformatics. 2023 Feb;21(1):108-126. doi: 10.1016/j.gpb.2022.02.006. Epub 2022 Mar 24. Genomics Proteomics Bioinformatics. 2023. PMID: 35341983 Free PMC article. Review.
Cited by
-
Research Progress and Application of Miniature CRISPR-Cas12 System in Gene Editing.Int J Mol Sci. 2024 Nov 26;25(23):12686. doi: 10.3390/ijms252312686. Int J Mol Sci. 2024. PMID: 39684395 Free PMC article. Review.
-
Molecular insights and rational engineering of a compact CRISPR-Cas effector Cas12h1 with a broad-spectrum PAM.Signal Transduct Target Ther. 2025 Feb 12;10(1):66. doi: 10.1038/s41392-025-02147-5. Signal Transduct Target Ther. 2025. PMID: 39955288 Free PMC article.
-
A New RNA-Dependent Cas12g Nuclease.Int J Mol Sci. 2023 Dec 4;24(23):17105. doi: 10.3390/ijms242317105. Int J Mol Sci. 2023. PMID: 38069429 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

