Discovery of neutralizing SARS-CoV-2 antibodies enriched in a unique antigen specific B cell cluster
- PMID: 37729215
- PMCID: PMC10511142
- DOI: 10.1371/journal.pone.0291131
Discovery of neutralizing SARS-CoV-2 antibodies enriched in a unique antigen specific B cell cluster
Abstract
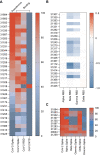
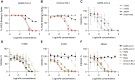
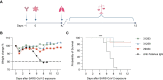
Despite development of effective SARS-CoV-2 vaccines, a sub-group of vaccine non-responders depends on therapeutic antibodies or small-molecule drugs in cases of severe disease. However, perpetual viral evolution has required continuous efficacy monitoring as well as exploration of new therapeutic antibodies, to circumvent resistance mutations arising in the viral population. We performed SARS-CoV-2-specific B cell sorting and subsequent single-cell sequencing on material from 15 SARS-CoV-2 convalescent participants. Through screening of 455 monoclonal antibodies for SARS-CoV-2 variant binding and virus neutralization, we identified a cluster of activated B cells highly enriched for SARS-CoV-2 neutralizing antibodies. Epitope binning and Cryo-EM structure analysis identified the majority of neutralizing antibodies having epitopes overlapping with the ACE2 receptor binding motif (class 1 binders). Extensive functional antibody characterization identified two potent neutralizing antibodies, one retaining SARS-CoV-1 neutralizing capability, while both bind major common variants of concern and display prophylactic efficacy in vivo. The transcriptomic signature of activated B cells harboring broadly binding neutralizing antibodies with therapeutic potential identified here, may be a guide in future efforts of rapid therapeutic antibody discovery.
Copyright: © 2023 Lende et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Structural Basis of a Human Neutralizing Antibody Specific to the SARS-CoV-2 Spike Protein Receptor-Binding Domain.Microbiol Spectr. 2021 Oct 31;9(2):e0135221. doi: 10.1128/Spectrum.01352-21. Epub 2021 Oct 13. Microbiol Spectr. 2021. PMID: 34643438 Free PMC article.
-
Broadly neutralizing SARS-CoV-2 antibodies through epitope-based selection from convalescent patients.Nat Commun. 2023 Feb 8;14(1):687. doi: 10.1038/s41467-023-36295-5. Nat Commun. 2023. PMID: 36755042 Free PMC article.
-
A Glycosylated RBD Protein Induces Enhanced Neutralizing Antibodies against Omicron and Other Variants with Improved Protection against SARS-CoV-2 Infection.J Virol. 2022 Sep 14;96(17):e0011822. doi: 10.1128/jvi.00118-22. Epub 2022 Aug 16. J Virol. 2022. PMID: 35972290 Free PMC article.
-
Identification of B-Cell Epitopes for Eliciting Neutralizing Antibodies against the SARS-CoV-2 Spike Protein through Bioinformatics and Monoclonal Antibody Targeting.Int J Mol Sci. 2022 Apr 14;23(8):4341. doi: 10.3390/ijms23084341. Int J Mol Sci. 2022. PMID: 35457159 Free PMC article. Review.
-
Microfluidic-assisted single-cell RNA sequencing facilitates the development of neutralizing monoclonal antibodies against SARS-CoV-2.Lab Chip. 2024 Feb 13;24(4):642-657. doi: 10.1039/d3lc00749a. Lab Chip. 2024. PMID: 38165771 Review.
Cited by
-
Monoclonal Antibody Generation Using Single B Cell Screening for Treating Infectious Diseases.BioDrugs. 2024 Jul;38(4):477-486. doi: 10.1007/s40259-024-00667-0. Epub 2024 Jul 2. BioDrugs. 2024. PMID: 38954386 Free PMC article. Review.
References
-
- Wei J, Stoesser N, Matthews PC, Ayoubkhani D, Studley R, Bell I, et al.. Antibody responses to SARS-CoV-2 vaccines in 45,965 adults from the general population of the United Kingdom. Nature microbiology. 2021;6(9):1140–9. Epub 2021/07/23. doi: 10.1038/s41564-021-00947-3 ; PubMed Central PMCID: PMC8294260 remaining authors declare no competing interests. - DOI - PMC - PubMed
-
- Søgaard OS, Reekie J, Johansen IS, Nielsen H, Benfield T, Wiese L, et al.. Characteristics associated with serological COVID-19 vaccine response and durability in an older population with significant comorbidity: the Danish Nationwide ENFORCE Study. Clinical microbiology and infection: the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2022;28(8):1126–33. Epub 2022/03/15. doi: 10.1016/j.cmi.2022.03.003 ; PubMed Central PMCID: PMC8913025. - DOI - PMC - PubMed
-
- Müller L, Andrée M, Moskorz W, Drexler I, Walotka L, Grothmann R, et al.. Age-dependent Immune Response to the Biontech/Pfizer BNT162b2 Coronavirus Disease 2019 Vaccination. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2021;73(11):2065–72. Epub 2021/04/28. doi: 10.1093/cid/ciab381 ; PubMed Central PMCID: PMC8135422. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

