Expanding the repertoire of human tandem repeat RNA-binding proteins
- PMID: 37729217
- PMCID: PMC10511089
- DOI: 10.1371/journal.pone.0290890
Expanding the repertoire of human tandem repeat RNA-binding proteins
Abstract
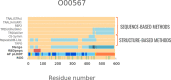
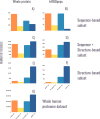
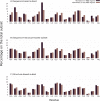
Protein regions consisting of arrays of tandem repeats are known to bind other molecular partners, including nucleic acid molecules. Although the interactions between repeat proteins and DNA are already widely explored, studies characterising tandem repeat RNA-binding proteins are lacking. We performed a large-scale analysis of human proteins devoted to expanding the knowledge about tandem repeat proteins experimentally reported as RNA-binding molecules. This work is timely because of the release of a full set of accurate structural models for the human proteome amenable to repeat detection using structural methods. The main goal of our analysis was to build a comprehensive set of human RNA-binding proteins that contain repeats at the sequence or structure level. Our results showed that the combination of sequence and structural methods finds significantly more tandem repeat proteins than either method alone. We identified 219 tandem repeat proteins that bind RNA molecules and characterised the overlap between repeat regions and RNA-binding regions as a first step towards assessing their functional relationship. We observed differences in the characteristics of repeat regions predicted by sequence-based or structure-based methods in terms of their sequence composition, their functions and their protein domains.
Copyright: © 2023 Ormazábal et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures











References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

