Apical PAR protein caps orient the mitotic spindle in C. elegans early embryos
- PMID: 37729910
- PMCID: PMC10615879
- DOI: 10.1016/j.cub.2023.08.069
Apical PAR protein caps orient the mitotic spindle in C. elegans early embryos
Abstract
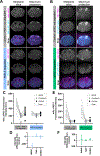
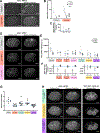
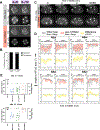
During embryonic development, oriented cell divisions are important for patterned tissue growth and cell fate specification. Cell division orientation is controlled in part by asymmetrically localized polarity proteins, which establish functional domains of the cell membrane and interact with microtubule regulators to position the mitotic spindle. For example, in the 8-cell mouse embryo, apical polarity proteins form caps on the outside, contact-free surface of the embryo that position the mitotic spindle to execute asymmetric cell division. A similar radial or "inside-outside" polarity is established at an early stage in many other animal embryos, but in most cases, it remains unclear how inside-outside polarity is established and how it influences downstream cell behaviors. Here, we explore inside-outside polarity in C. elegans somatic blastomeres using spatiotemporally controlled protein degradation and live embryo imaging. We show that PAR polarity proteins, which form apical caps at the center of the contact-free membrane, localize dynamically during the cell cycle and contribute to spindle orientation and proper cell positioning. Surprisingly, isolated single blastomeres lacking cell contacts are able to break symmetry and form PAR-3/atypical protein kinase C (aPKC) caps. Polarity caps form independently of actomyosin flows and microtubules and can regulate spindle orientation in cooperation with the key polarity kinase aPKC. Together, our results reveal a role for apical polarity caps in regulating spindle orientation in symmetrically dividing cells and provide novel insights into how these structures are formed.
Keywords: Caenorhabditis elegans; PAR proteins; cell polarity; embryo polarity; mitotic spindle orientation.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




Update of
-
Apical PAR-3 caps orient the mitotic spindle in C. elegans early embryos.bioRxiv [Preprint]. 2023 Mar 27:2023.03.27.534341. doi: 10.1101/2023.03.27.534341. bioRxiv. 2023. Update in: Curr Biol. 2023 Oct 23;33(20):4312-4329.e6. doi: 10.1016/j.cub.2023.08.069. PMID: 37034756 Free PMC article. Updated. Preprint.
Comment in
-
Cell polarity: Adapting the PAR cascade to diverse cellular contexts.Curr Biol. 2023 Oct 23;33(20):R1047-R1049. doi: 10.1016/j.cub.2023.08.083. Curr Biol. 2023. PMID: 37875077
Similar articles
-
Apical PAR-3 caps orient the mitotic spindle in C. elegans early embryos.bioRxiv [Preprint]. 2023 Mar 27:2023.03.27.534341. doi: 10.1101/2023.03.27.534341. bioRxiv. 2023. Update in: Curr Biol. 2023 Oct 23;33(20):4312-4329.e6. doi: 10.1016/j.cub.2023.08.069. PMID: 37034756 Free PMC article. Updated. Preprint.
-
The kinases PIG-1 and PAR-1 act in redundant pathways to regulate asymmetric division in the EMS blastomere of C. elegans.Dev Biol. 2018 Dec 1;444(1):9-19. doi: 10.1016/j.ydbio.2018.08.016. Epub 2018 Sep 10. Dev Biol. 2018. PMID: 30213539 Free PMC article.
-
PAR-3 and PAR-1 inhibit LET-99 localization to generate a cortical band important for spindle positioning in Caenorhabditis elegans embryos.Mol Biol Cell. 2007 Nov;18(11):4470-82. doi: 10.1091/mbc.e07-02-0105. Epub 2007 Aug 29. Mol Biol Cell. 2007. PMID: 17761536 Free PMC article.
-
Cell polarity and asymmetric cell division: the C. elegans early embryo.Essays Biochem. 2012;53:1-14. doi: 10.1042/bse0530001. Essays Biochem. 2012. PMID: 22928504 Review.
-
Par-3 family proteins in cell polarity & adhesion.FEBS J. 2022 Feb;289(3):596-613. doi: 10.1111/febs.15754. Epub 2021 Mar 3. FEBS J. 2022. PMID: 33565714 Free PMC article. Review.
Cited by
-
Origin and development of primary animal epithelia.Bioessays. 2024 Feb;46(2):e2300150. doi: 10.1002/bies.202300150. Epub 2023 Nov 27. Bioessays. 2024. PMID: 38009581 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

