Time-course transcriptome analysis of a double challenge bleomycin-induced lung fibrosis rat model uncovers ECM homoeostasis-related translationally relevant genes
- PMID: 37730279
- PMCID: PMC10510891
- DOI: 10.1136/bmjresp-2022-001476
Time-course transcriptome analysis of a double challenge bleomycin-induced lung fibrosis rat model uncovers ECM homoeostasis-related translationally relevant genes
Abstract
Background: Idiopathic pulmonary fibrosis (IPF) is an irreversible disorder with a poor prognosis. The incomplete understanding of IPF pathogenesis and the lack of accurate animal models is limiting the development of effective treatments. Thus, the selection of clinically relevant animal models endowed with similarities with the human disease in terms of lung anatomy, cell biology, pathways involved and genetics is essential. The bleomycin (BLM) intratracheal murine model is the most commonly used preclinical assay to evaluate new potential therapies for IPF. Here, we present the findings derived from an integrated histomorphometric and transcriptomic analysis to investigate the development of lung fibrosis in a time-course study in a BLM rat model and to evaluate its translational value in relation to IPF.
Methods: Rats were intratracheally injected with a double dose of BLM (days 0-4) and sacrificed at days 7, 14, 21, 28 and 56. Histomorphometric analysis of lung fibrosis was performed on left lung sections. Transcriptome profiling by RNAseq was performed on the right lung lobes and results were compared with nine independent human gene-expression IPF studies.
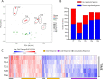
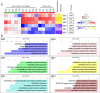
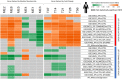
Results: The histomorphometric and transcriptomic analyses provided a detailed overview in terms of temporal gene-expression regulation during the establishment and repair of the fibrotic lesions. Moreover, the transcriptomic analysis identified three clusters of differentially coregulated genes whose expression was modulated in a time-dependent manner in response to BLM. One of these clusters, centred on extracellular matrix (ECM)-related process, was significantly correlated with histological parameters and gene sets derived from human IPF studies.
Conclusions: The model of lung fibrosis presented in this study lends itself as a valuable tool for preclinical efficacy evaluation of new potential drug candidates. The main finding was the identification of a group of persistently dysregulated genes, mostly related to ECM homoeostasis, which are shared with human IPF.
Keywords: Interstitial Fibrosis.
© Author(s) (or their employer(s)) 2023. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: VP, PC, SP, MGP, CM, GV, MC and MT are employees of Chiesi Farmaceutici S.p.A. FQ was engaged as consultant by Chiesi Farmaceutici S.p.A. All the remaining Authors have not actual and perceived conflicts of interest.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
