Integrated study of systemic and local airway transcriptomes in asthma reveals causal mediation of systemic effects by airway key drivers
- PMID: 37730635
- PMCID: PMC10512627
- DOI: 10.1186/s13073-023-01222-2
Integrated study of systemic and local airway transcriptomes in asthma reveals causal mediation of systemic effects by airway key drivers
Abstract
Background: Systemic and local profiles have each been associated with asthma, but parsing causal relationships between system-wide and airway-specific processes can be challenging. We sought to investigate systemic and airway processes in asthma and their causal relationships.
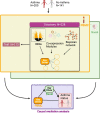
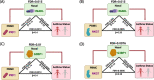
Methods: Three hundred forty-one participants with persistent asthma and non-asthmatic controls were recruited and underwent peripheral blood mononuclear cell (PBMC) collection and nasal brushing. Transcriptome-wide RNA sequencing of the PBMC and nasal samples and a series of analyses were then performed using a discovery and independent test set approach at each step to ensure rigor. Analytic steps included differential expression analyses, coexpression and probabilistic causal (Bayesian) network constructions, key driver analyses, and causal mediation models.
Results: Among the 341 participants, the median age was 13 years (IQR = 10-16), 164 (48%) were female, and 200 (58.7%) had persistent asthma with mean Asthma Control Test (ACT) score 16.6 (SD = 4.2). PBMC genes associated with asthma were enriched in co-expression modules for NK cell-mediated cytotoxicity (fold enrichment = 4.5, FDR = 6.47 × 10-32) and interleukin production (fold enrichment = 2.0, FDR = 1.01 × 10-15). Probabilistic causal network and key driver analyses identified NK cell granule protein (NKG7, fold change = 22.7, FDR = 1.02 × 10-31) and perforin (PRF1, fold change = 14.9, FDR = 1.31 × 10-22) as key drivers predicted to causally regulate PBMC asthma modules. Nasal genes associated with asthma were enriched in the tricarboxylic acid (TCA) cycle module (fold enrichment = 7.5 FDR = 5.09 × 10-107), with network analyses identifying G3BP stress granule assembly factor 1 (G3BP1, fold change = 9.1 FDR = 2.77 × 10-5) and InaD-like protein (INADL, fold change = 5.3 FDR = 2.98 × 10-9) as nasal key drivers. Causal mediation analyses revealed that associations between PBMC key drivers and asthma are causally mediated by nasal key drivers (FDR = 0.0076 to 0.015).
Conclusions: Integrated study of the systemic and airway transcriptomes in a well-phenotyped asthma cohort identified causal key drivers of asthma among PBMC and nasal transcripts. Associations between PBMC key drivers and asthma are causally mediated by nasal key drivers.
Keywords: Airway; Asthma; Blood; Causal network; Interleukin; NK cell; Nasal; Peripheral blood mononuclear cell; Transcriptome; Tricarboxylic acid.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


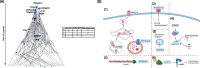
Similar articles
-
Joint transcriptomic and cytometric study of children with peanut allergy reveals molecular and cellular cross talk in reaction thresholds.J Allergy Clin Immunol. 2024 Jun;153(6):1721-1728. doi: 10.1016/j.jaci.2023.12.028. Epub 2024 Jan 23. J Allergy Clin Immunol. 2024. PMID: 38272374 Free PMC article.
-
Network study of nasal transcriptome profiles reveals master regulator genes of asthma.J Allergy Clin Immunol. 2021 Mar;147(3):879-893. doi: 10.1016/j.jaci.2020.07.006. Epub 2020 Aug 20. J Allergy Clin Immunol. 2021. PMID: 32828590 Free PMC article.
-
Dissecting childhood asthma with nasal transcriptomics distinguishes subphenotypes of disease.J Allergy Clin Immunol. 2014 Mar;133(3):670-8.e12. doi: 10.1016/j.jaci.2013.11.025. Epub 2014 Feb 2. J Allergy Clin Immunol. 2014. PMID: 24495433 Free PMC article.
-
Impaired type I interferon regulation in the blood transcriptome of recurrent asthma exacerbations.BMC Med Genomics. 2018 Feb 27;11(1):21. doi: 10.1186/s12920-018-0340-3. BMC Med Genomics. 2018. PMID: 29486764 Free PMC article.
-
Identification of surrogate prognostic biomarkers for allergic asthma in nasal epithelial brushing samples by WGCNA.J Cell Biochem. 2019 Apr;120(4):5137-5150. doi: 10.1002/jcb.27790. Epub 2018 Oct 10. J Cell Biochem. 2019. PMID: 30304558
Cited by
-
Lessons from national biobank projects utilizing whole-genome sequencing for population-scale genomics.Genomics Inform. 2025 Mar 6;23(1):8. doi: 10.1186/s44342-025-00040-9. Genomics Inform. 2025. PMID: 40050991 Free PMC article. Review.
-
Leveraging baseline transcriptional features and information from single-cell data to power the prediction of influenza vaccine response.Front Cell Infect Microbiol. 2024 Feb 7;14:1243586. doi: 10.3389/fcimb.2024.1243586. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38384303 Free PMC article.
-
Analytical challenges in omics research on asthma and allergy: A National Institute of Allergy and Infectious Diseases workshop.J Allergy Clin Immunol. 2024 Apr;153(4):954-968. doi: 10.1016/j.jaci.2024.01.014. Epub 2024 Jan 29. J Allergy Clin Immunol. 2024. PMID: 38295882 Free PMC article.
References
-
- Asher MI, Garcia-Marcos L, Pearce NE, Strachan DP. Trends in worldwide asthma prevalence. Eur Respir J. 2020;56:2002094. - PubMed
-
- Porsbjerg C, Melen E, Lehtimaki L, Shaw D. Asthma. Lancet. 2023;401:858–73. - PubMed
-
- Edwards MR, Saglani S, Schwarze J, Skevaki C, Smith JA, Ainsworth B, et al. Addressing unmet needs in understanding asthma mechanisms: From the European Asthma Research and Innovation Partnership (EARIP) Work Package (WP)2 collaborators. Eur Respir J. 2017;49:1602448. - PubMed
-
- Gauvreau GM, El-Gammal AI, O’Byrne PM. Allergen-induced airway responses. Eur Respir J. 2015;46:819–831. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

