Critical requirement of SOS1 for tumor development and microenvironment modulation in KRASG12D-driven lung adenocarcinoma
- PMID: 37730692
- PMCID: PMC10511506
- DOI: 10.1038/s41467-023-41583-1
Critical requirement of SOS1 for tumor development and microenvironment modulation in KRASG12D-driven lung adenocarcinoma
Abstract
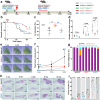
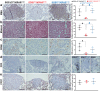
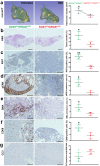
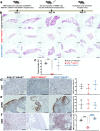
The impact of genetic ablation of SOS1 or SOS2 is evaluated in a murine model of KRASG12D-driven lung adenocarcinoma (LUAD). SOS2 ablation shows some protection during early stages but only SOS1 ablation causes significant, specific long term increase of survival/lifespan of the KRASG12D mice associated to markedly reduced tumor burden and reduced populations of cancer-associated fibroblasts, macrophages and T-lymphocytes in the lung tumor microenvironment (TME). SOS1 ablation also causes specific shrinkage and regression of LUAD tumoral masses and components of the TME in pre-established KRASG12D LUAD tumors. The critical requirement of SOS1 for KRASG12D-driven LUAD is further confirmed by means of intravenous tail injection of KRASG12D tumor cells into SOS1KO/KRASWT mice, or of SOS1-less, KRASG12D tumor cells into wildtype mice. In silico analyses of human lung cancer databases support also the dominant role of SOS1 regarding tumor development and survival in LUAD patients. Our data indicate that SOS1 is critically required for development of KRASG12D-driven LUAD and confirm the validity of this RAS-GEF activator as an actionable therapeutic target in KRAS mutant LUAD.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

