NET4 and RabG3 link actin to the tonoplast and facilitate cytoskeletal remodelling during stomatal immunity
- PMID: 37730720
- PMCID: PMC10511709
- DOI: 10.1038/s41467-023-41337-z
NET4 and RabG3 link actin to the tonoplast and facilitate cytoskeletal remodelling during stomatal immunity
Abstract
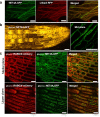
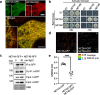
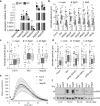
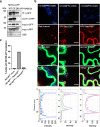
Members of the NETWORKED (NET) family are involved in actin-membrane interactions. Here we show that two members of the NET family, NET4A and NET4B, are essential for normal guard cell actin reorganization, which is a process critical for stomatal closure in plant immunity. NET4 proteins interact with F-actin and with members of the Rab7 GTPase RABG3 family through two distinct domains, allowing for simultaneous localization to actin filaments and the tonoplast. NET4 proteins interact with GTP-bound, active RABG3 members, suggesting their function being downstream effectors. We also show that RABG3b is critical for stomatal closure induced by microbial patterns. Taken together, we conclude that the actin cytoskeletal remodelling during stomatal closure involves a molecular link between actin filaments and the tonoplast, which is mediated by the NET4-RABG3b interaction. We propose that stomatal closure to microbial patterns involves the coordinated action of immune-triggered osmotic changes and actin cytoskeletal remodelling likely driving compact vacuolar morphologies.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors have no competing interests.
Figures


Similar articles
-
Vacuolar occupancy is crucial for cell elongation and growth regardless of the underlying mechanism.Plant Signal Behav. 2021 Aug 3;16(8):1922796. doi: 10.1080/15592324.2021.1922796. Epub 2021 May 3. Plant Signal Behav. 2021. PMID: 33938395 Free PMC article.
-
Dynamics of vacuoles and actin filaments in guard cells and their roles in stomatal movement.Plant Cell Environ. 2009 Aug;32(8):1108-16. doi: 10.1111/j.1365-3040.2009.01993.x. Epub 2009 Apr 22. Plant Cell Environ. 2009. PMID: 19422610 Review.
-
The reorganization of actin filaments is required for vacuolar fusion of guard cells during stomatal opening in Arabidopsis.Plant Cell Environ. 2013 Feb;36(2):484-97. doi: 10.1111/j.1365-3040.2012.02592.x. Epub 2012 Sep 13. Plant Cell Environ. 2013. PMID: 22891733
-
AP3M harbors actin filament binding activity that is crucial for vacuole morphology and stomatal closure in Arabidopsis.Proc Natl Acad Sci U S A. 2019 Sep 3;116(36):18132-18141. doi: 10.1073/pnas.1901431116. Epub 2019 Aug 20. Proc Natl Acad Sci U S A. 2019. PMID: 31431522 Free PMC article.
-
Ras-related GTPases and the cytoskeleton.Mol Biol Cell. 1992 May;3(5):475-9. doi: 10.1091/mbc.3.5.475. Mol Biol Cell. 1992. PMID: 1611153 Free PMC article. Review.
Cited by
-
Plant NETWORKED and VAP27 Proteins Work in Complexes to Regulate Membrane-Based Functions.Contact (Thousand Oaks). 2025 May 27;8:25152564251342533. doi: 10.1177/25152564251342533. eCollection 2025 Jan-Dec. Contact (Thousand Oaks). 2025. PMID: 40458073 Free PMC article. Review.
-
Evolution and subfunctionalization of CIPK6 homologous genes in regulating cotton drought resistance.Nat Commun. 2024 Jul 9;15(1):5733. doi: 10.1038/s41467-024-50097-3. Nat Commun. 2024. PMID: 38977687 Free PMC article.
References
-
- Deeks MJ, et al. A superfamily of actin-binding proteins at the actin-membrane nexus of higher plants. Curr. Biol. 2012;22:1595–1600. - PubMed
-
- Wang P, Hawkins TJ, Hussey PJ. Connecting membranes to the actin cytoskeleton. Curr. Opin. Plant Biol. 2017;40:71–76. - PubMed
-
- Duckney P, et al. Actin-membrane interactions mediated by NETWORKED2 in Arabidopsis pollen tubes through associations with Pollen Receptor-Like Kinase 4 and 5. New Phytol. 2017;216:1170–1180. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

