A subset of viruses thrives following microbial resuscitation during rewetting of a seasonally dry California grassland soil
- PMID: 37730729
- PMCID: PMC10511743
- DOI: 10.1038/s41467-023-40835-4
A subset of viruses thrives following microbial resuscitation during rewetting of a seasonally dry California grassland soil
Abstract
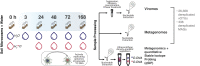
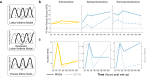
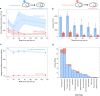
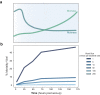
Viruses are abundant, ubiquitous members of soil communities that kill microbial cells, but how they respond to perturbation of soil ecosystems is essentially unknown. Here, we investigate lineage-specific virus-host dynamics in grassland soil following "wet-up", when resident microbes are both resuscitated and lysed after a prolonged dry period. Quantitative isotope tracing, time-resolved metagenomics and viromic analyses indicate that dry soil holds a diverse but low biomass reservoir of virions, of which only a subset thrives following wet-up. Viral richness decreases by 50% within 24 h post wet-up, while viral biomass increases four-fold within one week. Though recent hypotheses suggest lysogeny predominates in soil, our evidence indicates that viruses in lytic cycles dominate the response to wet-up. We estimate that viruses drive a measurable and continuous rate of cell lysis, with up to 46% of microbial death driven by viral lysis one week following wet-up. Thus, viruses contribute to turnover of soil microbial biomass and the widely reported CO2 efflux following wet-up of seasonally dry soils.
© 2023. Springer Nature Limited.
Conflict of interest statement
J.F.B. is a founder of Metagenomi. The remaining authors declare no competing interests.
Figures

References
-
- Williamson KE, Fuhrmann JJ, Wommack KE, Radosevich M. Viruses in soil ecosystems: an unknown quantity within an unexplored territory. Annu. Rev. Virol. 2017;4:201–219. - PubMed
-
- Kuzyakov Y, Mason-Jones K. Viruses in soil: nano-scale undead drivers of microbial life, biogeochemical turnover and ecosystem functions. Soil Biol. Biochem. 2018;127:305–317.
-
- Paez-Espino, D. et al. Uncovering Earth’s Virome. Nature10.1038/nature19094 (2016). - PubMed
-
- Pratama AA, van Elsas JD. The ‘Neglected’ soil virome—potential role and impact. Trends Microbiol. 2018;26:649–662. - PubMed

