Cryo-EM structures reveal native GABAA receptor assemblies and pharmacology
- PMID: 37730991
- PMCID: PMC10550821
- DOI: 10.1038/s41586-023-06556-w
Cryo-EM structures reveal native GABAA receptor assemblies and pharmacology
Abstract
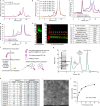
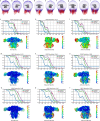
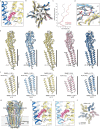
Type A γ-aminobutyric acid receptors (GABAARs) are the principal inhibitory receptors in the brain and the target of a wide range of clinical agents, including anaesthetics, sedatives, hypnotics and antidepressants1-3. However, our understanding of GABAAR pharmacology has been hindered by the vast number of pentameric assemblies that can be derived from 19 different subunits4 and the lack of structural knowledge of clinically relevant receptors. Here, we isolate native murine GABAAR assemblies containing the widely expressed α1 subunit and elucidate their structures in complex with drugs used to treat insomnia (zolpidem (ZOL) and flurazepam) and postpartum depression (the neurosteroid allopregnanolone (APG)). Using cryo-electron microscopy (cryo-EM) analysis and single-molecule photobleaching experiments, we uncover three major structural populations in the brain: the canonical α1β2γ2 receptor containing two α1 subunits, and two assemblies containing one α1 and either an α2 or α3 subunit, in which the single α1-containing receptors feature a more compact arrangement between the transmembrane and extracellular domains. Interestingly, APG is bound at the transmembrane α/β subunit interface, even when not added to the sample, revealing an important role for endogenous neurosteroids in modulating native GABAARs. Together with structurally engaged lipids, neurosteroids produce global conformational changes throughout the receptor that modify the ion channel pore and the binding sites for GABA and insomnia medications. Our data reveal the major α1-containing GABAAR assemblies, bound with endogenous neurosteroid, thus defining a structural landscape from which subtype-specific drugs can be developed.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures










Comment in
-
Understanding GABAA receptor pharmacology.Nat Rev Drug Discov. 2023 Nov;22(11):873. doi: 10.1038/d41573-023-00162-1. Nat Rev Drug Discov. 2023. PMID: 37798464 No abstract available.
References
-
- Barnard EA, et al. International Union of Pharmacology. XV. Subtypes of γ-aminobutyric acidA receptors: classification on the basis of subunit structure and receptor function. Pharmacol. Rev. 1998;50:291–313. - PubMed
-
- Nemeroff CB. The role of GABA in the pathophysiology and treatment of anxiety disorders. Psychopharmacol. Bull. 2003;37:133–146. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

