The β1-adrenergic receptor links sympathetic nerves to T cell exhaustion
- PMID: 37731001
- PMCID: PMC10871066
- DOI: 10.1038/s41586-023-06568-6
The β1-adrenergic receptor links sympathetic nerves to T cell exhaustion
Abstract
CD8+ T cells are essential components of the immune response against viral infections and tumours, and are capable of eliminating infected and cancerous cells. However, when the antigen cannot be cleared, T cells enter a state known as exhaustion1. Although it is clear that chronic antigen contributes to CD8+ T cell exhaustion, less is known about how stress responses in tissues regulate T cell function. Here we show a new link between the stress-associated catecholamines and the progression of T cell exhaustion through the β1-adrenergic receptor ADRB1. We identify that exhausted CD8+ T cells increase ADRB1 expression and that exposure of ADRB1+ T cells to catecholamines suppresses their cytokine production and proliferation. Exhausted CD8+ T cells cluster around sympathetic nerves in an ADRB1-dependent manner. Ablation of β1-adrenergic signalling limits the progression of T cells towards the exhausted state in chronic infection and improves effector functions when combined with immune checkpoint blockade (ICB) in melanoma. In a pancreatic cancer model resistant to ICB, β-blockers and ICB synergize to boost CD8+ T cell responses and induce the development of tissue-resident memory-like T cells. Malignant disease is associated with increased catecholamine levels in patients2,3, and our results establish a connection between the sympathetic stress response, tissue innervation and T cell exhaustion. Here, we uncover a new mechanism by which blocking β-adrenergic signalling in CD8+ T cells rejuvenates anti-tumour functions.
© 2023. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Figures



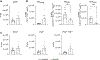


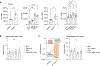








Comment in
-
Stress can be exhausting for T cells.Nat Rev Drug Discov. 2023 Nov;22(11):870. doi: 10.1038/d41573-023-00157-y. Nat Rev Drug Discov. 2023. PMID: 37783811 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

