This is a preprint.
Pervasive mislocalization of pathogenic coding variants underlying human disorders
- PMID: 37732209
- PMCID: PMC10508771
- DOI: 10.1101/2023.09.05.556368
Pervasive mislocalization of pathogenic coding variants underlying human disorders
Update in
-
Pervasive mislocalization of pathogenic coding variants underlying human disorders.Cell. 2024 Nov 14;187(23):6725-6741.e13. doi: 10.1016/j.cell.2024.09.003. Epub 2024 Sep 30. Cell. 2024. PMID: 39353438
Abstract
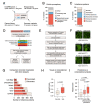
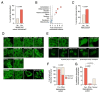
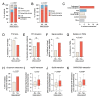
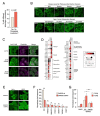
Widespread sequencing has yielded thousands of missense variants predicted or confirmed as disease-causing. This creates a new bottleneck: determining the functional impact of each variant - largely a painstaking, customized process undertaken one or a few genes or variants at a time. Here, we established a high-throughput imaging platform to assay the impact of coding variation on protein localization, evaluating 3,547 missense variants of over 1,000 genes and phenotypes. We discovered that mislocalization is a common consequence of coding variation, affecting about one-sixth of all pathogenic missense variants, all cellular compartments, and recessive and dominant disorders alike. Mislocalization is primarily driven by effects on protein stability and membrane insertion rather than disruptions of trafficking signals or specific interactions. Furthermore, mislocalization patterns help explain pleiotropy and disease severity and provide insights on variants of unknown significance. Our publicly available resource will likely accelerate the understanding of coding variation in human diseases.
Conflict of interest statement
DECLARATION OF INTERESTS A.E.C. serves as a scientific advisor for Recursion, which uses image-based profiling and Cell Painting for drug discovery and receives honoraria for occasional talks at pharmaceutical and biotechnology companies. The other authors declare no competing interests relevant to this manuscript.
Figures

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
