This is a preprint.
The Fanconi anemia core complex promotes CtIP-dependent end-resection to drive homologous recombination at DNA double-strand breaks
- PMID: 37732274
- PMCID: PMC10508776
- DOI: 10.1101/2023.09.05.556391
The Fanconi anemia core complex promotes CtIP-dependent end-resection to drive homologous recombination at DNA double-strand breaks
Update in
-
The Fanconi anemia core complex promotes CtIP-dependent end resection to drive homologous recombination at DNA double-strand breaks.Nat Commun. 2024 Aug 16;15(1):7076. doi: 10.1038/s41467-024-51090-6. Nat Commun. 2024. PMID: 39152113 Free PMC article.
Abstract
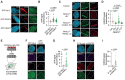
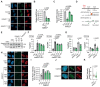
Homologous Recombination (HR) is a high-fidelity repair mechanism of DNA Double-Strand Breaks (DSBs), which are induced by irradiation, genotoxic chemicals or physiological DNA damaging processes. DSBs are also generated as intermediates during the repair of interstrand crosslinks (ICLs). In this context, the Fanconi anemia (FA) core complex, which is effectively recruited to ICLs, promotes HR-mediated DSB-repair. However, whether the FA core complex also promotes HR at ICL-independent DSBs remains controversial. Here, we identified the FA core complex members FANCL and Ube2T as HR-promoting factors in a CRISPR/Cas9-based screen with cells carrying the DSB-repair reporter DSB-Spectrum. Using isogenic cell-line models, we validated the HR-function of FANCL and Ube2T, and demonstrated a similar function for their ubiquitination-substrate FANCD2. We further show that FANCL and Ube2T are directly recruited to DSBs and are required for the accumulation of FANCD2 at these break sites. Mechanistically, we demonstrate that FANCL ubiquitin ligase activity is required for the accumulation of the nuclease CtIP at DSBs, and consequently for optimal end-resection and Rad51 loading. CtIP overexpression rescues HR in FANCL-deficient cells, validating that FANCL primarily regulates HR by promoting CtIP recruitment. Together, these data demonstrate that the FA core complex and FANCD2 have a dual genome maintenance function by promoting repair of DSBs as well as the repair of ICLs.
Figures




References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
