The role of integration host factor in biofilm and virulence of high-alcohol-producing Klebsiella pneumoniae
- PMID: 37732783
- PMCID: PMC10581059
- DOI: 10.1128/spectrum.01170-23
The role of integration host factor in biofilm and virulence of high-alcohol-producing Klebsiella pneumoniae
Abstract
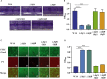
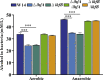
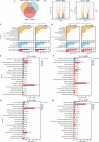
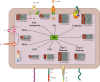
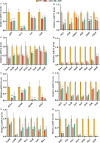
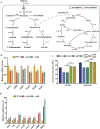
Klebsiella pneumoniae is a well-known human nosocomial pathogen with an arsenal of virulence factors, including capsular polysaccharides (CPS), fimbriae, flagella, and lipopolysaccharides (LPS). Our previous study found that alcohol acted as an essential virulence factor for high-alcohol-producing K. pneumoniae (HiAlc Kpn). Integration host factor (IHF) is a nucleoid-associated protein that functions as a global virulence regulator in Escherichia coli. However, the regulatory role of IHF in K. pneumoniae remains unknown. In the present study, we found that deletion of ihfA or ihfB resulted in a slight defect in bacterial growth, a severe absence of biofilm formation and cytotoxicity, and a significant reduction in alcohol production. RNA sequencing differential gene expression analysis showed that compared with the wild-type control, the expression of many virulence factor genes was downregulated in ΔihfA and ΔihfB strains, such as those related to CPS (rcsA, galF, wzi, and iscR), LPS (rfbABCD), type I and type III fimbriae (fim and mrk operon), cellulose (bcs operon), iron transporter (feoABC, fhuA, fhuF, tonB, exbB, and exbD), quorum sensing (lsr operon and sdiA), type II secretion system (T2SS) and type VI secretion system (T6SS) (tssG, hcp, and gspE). Of these virulence factors, CPS, LPS, fimbriae, and cellulose are involved in biofilm formation. In addition, IHF could affect the alcohol production by regulating genes related to glucose intake (ptsG), pyruvate formate-lyase, alcohol dehydrogenase, and the tricarboxylic acid (TCA) cycle. Our data provided new insights into the importance of IHF in regulating the virulence of HiAlc Kpn. IMPORTANCE Klebsiella pneumoniae is a well-known human nosocomial pathogen that causes various infectious diseases, including urinary tract infections, hospital-acquired pneumonia, bacteremia, and liver abscesses. Our previous studies demonstrated that HiAlc Kpn mediated the development of nonalcoholic fatty liver disease by producing excess endogenous alcohol in vivo. However, the regulators regulating the expression of genes related to metabolism, biofilm formation, and virulence of HiAlc Kpn remain unclear. In this study, the regulator IHF was found to positively regulate biofilm formation and many virulence factors including CPS, LPS, type I and type III fimbriae, cellulose, iron transporter, AI-2 quorum sensing, T2SS, and T6SS in HiAlc Kpn. Furthermore, IHF positively regulated alcohol production in HiAlc Kpn. Our results suggested that IHF could be a potential drug target for treating various infectious diseases caused by K. pneumoniae. Hence, the regulation of different virulence factors by IHF in K. pneumoniae requires further investigation.
Keywords: HiAlc Kpn; IHF; biofilm; regulate; virulence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Yuan J, Chen C, Cui J, Lu J, Yan C, Wei X, Zhao X, Li N, Li S, Xue G, Cheng W, Li B, Li H, Lin W, Tian C, Zhao J, Han J, An D, Zhang Q, Wei H, Zheng M, Ma X, Li W, Chen X, Zhang Z, Zeng H, Ying S, Wu J, Yang R, Liu D. 2019. Fatty liver disease caused by high-alcohol-producing Klebsiella pneumoniae. Cell Metab 30:1172. doi:10.1016/j.cmet.2019.11.006 - DOI - PubMed
-
- Federici S, Kredo-Russo S, Valdés-Mas R, Kviatcovsky D, Weinstock E, Matiuhin Y, Silberberg Y, Atarashi K, Furuichi M, Oka A, Liu B, Fibelman M, Weiner IN, Khabra E, Cullin N, Ben-Yishai N, Inbar D, Ben-David H, Nicenboim J, Kowalsman N, Lieb W, Kario E, Cohen T, Geffen YF, Zelcbuch L, Cohen A, Rappo U, Gahali-Sass I, Golembo M, Lev V, Dori-Bachash M, Shapiro H, Moresi C, Cuevas-Sierra A, Mohapatra G, Kern L, Zheng D, Nobs SP, Suez J, Stettner N, Harmelin A, Zak N, Puttagunta S, Bassan M, Honda K, Sokol H, Bang C, Franke A, Schramm C, Maharshak N, Sartor RB, Sorek R, Elinav E. 2022. Targeted suppression of human ibd-associated gut microbiota commensals by phage consortia for treatment of intestinal inflammation. Cell 185:2879–2898. doi:10.1016/j.cell.2022.07.003 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

