PKR-mediated stress response enhances dengue and Zika virus replication
- PMID: 37732809
- PMCID: PMC10653888
- DOI: 10.1128/mbio.00934-23
PKR-mediated stress response enhances dengue and Zika virus replication
Abstract
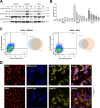
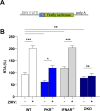
One of the fundamental features that make viruses intracellular parasites is the necessity to use cellular translational machinery. Hence, this is a crucial checkpoint for controlling infections. Here, we show that dengue and Zika viruses, responsible for nearly 400 million infections every year worldwide, explore such control for optimal replication. Using immunocompetent cells, we demonstrate that arrest of protein translations happens after sensing of dsRNA and that the information required to avoid this blocking is contained in viral 5'-UTR. Our work, therefore, suggests that the non-canonical translation described for these viruses is engaged when the intracellular stress response is activated.
Keywords: PKR; Zika virus; dengue virus; innate immunity; protein translation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
IFN alpha inducible protein 27 (IFI27) acts as a positive regulator of PACT-dependent PKR activation after RNA virus infections.PLoS Pathog. 2025 Jun 16;21(6):e1013246. doi: 10.1371/journal.ppat.1013246. eCollection 2025 Jun. PLoS Pathog. 2025. PMID: 40522981 Free PMC article.
-
Replication in the presence of dengue convalescent serum impacts Zika virus neutralization sensitivity and fitness.Front Cell Infect Microbiol. 2023 Mar 9;13:1130749. doi: 10.3389/fcimb.2023.1130749. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 36968111 Free PMC article.
-
CMPK2 restricts Zika virus replication by inhibiting viral translation.PLoS Pathog. 2023 Apr 19;19(4):e1011286. doi: 10.1371/journal.ppat.1011286. eCollection 2023 Apr. PLoS Pathog. 2023. PMID: 37075076 Free PMC article.
-
Clinical manifestations of dengue, Zika and chikungunya in the Pacific Islands: A systematic review and meta-analysis.Rev Med Virol. 2024 Mar;34(2):e2521. doi: 10.1002/rmv.2521. Rev Med Virol. 2024. PMID: 38340071
-
Congenital Zika syndrome: Pitfalls in the placental barrier.Rev Med Virol. 2018 Sep;28(5):e1985. doi: 10.1002/rmv.1985. Epub 2018 May 15. Rev Med Virol. 2018. PMID: 29761581
Cited by
-
APOBEC3B drives PKR-mediated translation shutdown and protects stress granules in response to viral infection.Nat Commun. 2023 Feb 14;14(1):820. doi: 10.1038/s41467-023-36445-9. Nat Commun. 2023. PMID: 36781883 Free PMC article.
-
Zika virus non-coding RNAs antagonize antiviral responses by PKR-mediated translational arrest.Nucleic Acids Res. 2024 Oct 14;52(18):11128-11147. doi: 10.1093/nar/gkae507. Nucleic Acids Res. 2024. PMID: 38917323 Free PMC article.
-
Cooperative role of PACT and ADAR1 in preventing aberrant PKR activation by self-derived double-stranded RNA.Nat Commun. 2025 Apr 5;16(1):3246. doi: 10.1038/s41467-025-58412-2. Nat Commun. 2025. PMID: 40185749 Free PMC article.
-
Activation of ATF3 via the integrated stress response pathway regulates innate immune response to restrict Zika virus.J Virol. 2024 Oct 22;98(10):e0105524. doi: 10.1128/jvi.01055-24. Epub 2024 Aug 30. J Virol. 2024. PMID: 39212382 Free PMC article.
-
Zika viruses encode 5' upstream open reading frames affecting infection of human brain cells.Nat Commun. 2024 Oct 12;15(1):8822. doi: 10.1038/s41467-024-53085-9. Nat Commun. 2024. PMID: 39394194 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

