A paracrine circuit of IL-1β/IL-1R1 between myeloid and tumor cells drives genotype-dependent glioblastoma progression
- PMID: 37733448
- PMCID: PMC10645395
- DOI: 10.1172/JCI163802
A paracrine circuit of IL-1β/IL-1R1 between myeloid and tumor cells drives genotype-dependent glioblastoma progression
Abstract
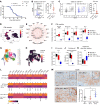
Monocytes and monocyte-derived macrophages (MDMs) from blood circulation infiltrate glioblastoma (GBM) and promote growth. Here, we show that PDGFB-driven GBM cells induce the expression of the potent proinflammatory cytokine IL-1β in MDM, which engages IL-1R1 in tumor cells, activates the NF-κB pathway, and subsequently leads to induction of monocyte chemoattractant proteins (MCPs). Thus, a feedforward paracrine circuit of IL-1β/IL-1R1 between tumors and MDM creates an interdependence driving PDGFB-driven GBM progression. Genetic loss or locally antagonizing IL-1β/IL-1R1 leads to reduced MDM infiltration, diminished tumor growth, and reduced exhausted CD8+ T cells and thereby extends the survival of tumor-bearing mice. In contrast to IL-1β, IL-1α exhibits antitumor effects. Genetic deletion of Il1a/b is associated with decreased recruitment of lymphoid cells and loss-of-interferon signaling in various immune populations and subsets of malignant cells and is associated with decreased survival time of PDGFB-driven tumor-bearing mice. In contrast to PDGFB-driven GBM, Nf1-silenced tumors have a constitutively active NF-κB pathway, which drives the expression of MCPs to recruit monocytes into tumors. These results indicate local antagonism of IL-1β could be considered as an effective therapy specifically for proneural GBM.
Keywords: Brain cancer; Cancer immunotherapy; Immunology; Macrophages; Oncology.
Figures








Comment in
-
Breaking the feed forward inflammatory cytokine loop in the tumor microenvironment of PDGFB-driven glioblastomas.J Clin Invest. 2023 Nov 15;133(22):e175127. doi: 10.1172/JCI175127. J Clin Invest. 2023. PMID: 37966120 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

