Susceptibility to Zika virus in a Collaborative Cross mouse strain is induced by Irf3 deficiency in vitro but requires other variants in vivo
- PMID: 37733807
- PMCID: PMC10547207
- DOI: 10.1371/journal.ppat.1011446
Susceptibility to Zika virus in a Collaborative Cross mouse strain is induced by Irf3 deficiency in vitro but requires other variants in vivo
Abstract
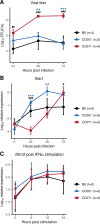
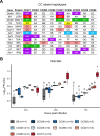
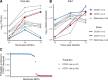
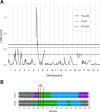
Zika virus (ZIKV) is a Flavivirus responsible for recent epidemics in Pacific Islands and in the Americas. In humans, the consequences of ZIKV infection range from asymptomatic infection to severe neurological disease such as Guillain-Barré syndrome or fetal neurodevelopmental defects, suggesting, among other factors, the influence of host genetic variants. We previously reported similar diverse outcomes of ZIKV infection in mice of the Collaborative Cross (CC), a collection of inbred strains with large genetic diversity. CC071/TauUnc (CC071) was the most susceptible CC strain with severe symptoms and lethality. Notably, CC071 has been recently reported to be also susceptible to other flaviviruses including dengue virus, Powassan virus, West Nile virus, and to Rift Valley fever virus. To identify the genetic origin of this broad susceptibility, we investigated ZIKV replication in mouse embryonic fibroblasts (MEFs) from CC071 and two resistant strains. CC071 showed uncontrolled ZIKV replication associated with delayed induction of type-I interferons (IFN-I). Genetic analysis identified a mutation in the Irf3 gene specific to the CC071 strain which prevents the protein phosphorylation required to activate interferon beta transcription. We demonstrated that this mutation induces the same defective IFN-I response and uncontrolled viral replication in MEFs as an Irf3 knock-out allele. By contrast, we also showed that Irf3 deficiency did not induce the high plasma viral load and clinical severity observed in CC071 mice and that susceptibility alleles at other genes, not associated with the IFN-I response, are required. Our results provide new insight into the in vitro and in vivo roles of Irf3, and into the genetic complexity of host responses to flaviviruses.
Copyright: © 2023 Bourdon et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Genetic Diversity of Collaborative Cross Mice Controls Viral Replication, Clinical Severity, and Brain Pathology Induced by Zika Virus Infection, Independently of Oas1b.J Virol. 2020 Jan 17;94(3):e01034-19. doi: 10.1128/JVI.01034-19. Print 2020 Jan 17. J Virol. 2020. PMID: 31694939 Free PMC article.
-
Neuroinvasive Flavivirus Pathogenesis Is Restricted by Host Genetic Factors in Collaborative Cross Mice, Independently of Oas1b.J Virol. 2023 Jul 27;97(7):e0071523. doi: 10.1128/jvi.00715-23. Epub 2023 Jun 13. J Virol. 2023. PMID: 37310228 Free PMC article.
-
Zika virus infection of mature neurons from immunocompetent mice generates a disease-associated microglia and a tauopathy-like phenotype in link with a delayed interferon beta response.J Neuroinflammation. 2022 Dec 20;19(1):307. doi: 10.1186/s12974-022-02668-8. J Neuroinflammation. 2022. PMID: 36539803 Free PMC article.
-
Hide and Seek: The Interplay Between Zika Virus and the Host Immune Response.Front Immunol. 2021 Oct 21;12:750365. doi: 10.3389/fimmu.2021.750365. eCollection 2021. Front Immunol. 2021. PMID: 34745123 Free PMC article. Review.
-
Functional RNA during Zika virus infection.Virus Res. 2018 Aug 2;254:41-53. doi: 10.1016/j.virusres.2017.08.015. Epub 2017 Aug 31. Virus Res. 2018. PMID: 28864425 Review.
Cited by
-
Protein kinase R induced by type I interferons is a main regulator of reactive microglia in Zika virus infection.Glia. 2025 Jan;73(1):80-104. doi: 10.1002/glia.24619. Epub 2024 Oct 3. Glia. 2025. PMID: 39359232 Free PMC article.
-
Intranasal Immunization for Zika in a Pre-Clinical Model.Viruses. 2024 May 28;16(6):865. doi: 10.3390/v16060865. Viruses. 2024. PMID: 38932158 Free PMC article.
References
-
- Oliveira Melo AS, Malinger G, Ximenes R, Szejnfeld PO, Alves Sampaio S, Bispo de Filippis AM. Zika virus intrauterine infection causes fetal brain abnormality and microcephaly: tip of the iceberg?: Physician Alert. Ultrasound Obstet Gynecol. janv 2016;47(1):6–7. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

