A bioinformatics screen reveals hox and chromatin remodeling factors at the Drosophila histone locus
- PMID: 37735352
- PMCID: PMC10515271
- DOI: 10.1186/s12863-023-01147-0
A bioinformatics screen reveals hox and chromatin remodeling factors at the Drosophila histone locus
Abstract
Background: Cells orchestrate histone biogenesis with strict temporal and quantitative control. To efficiently regulate histone biogenesis, the repetitive Drosophila melanogaster replication-dependent histone genes are arrayed and clustered at a single locus. Regulatory factors concentrate in a nuclear body known as the histone locus body (HLB), which forms around the locus. Historically, HLB factors are largely discovered by chance, and few are known to interact directly with DNA. It is therefore unclear how the histone genes are specifically targeted for unique and coordinated regulation.
Results: To expand the list of known HLB factors, we performed a candidate-based screen by mapping 30 publicly available ChIP datasets of 27 unique factors to the Drosophila histone gene array. We identified novel transcription factor candidates, including the Drosophila Hox proteins Ultrabithorax (Ubx), Abdominal-A (Abd-A), and Abdominal-B (Abd-B), suggesting a new pathway for these factors in influencing body plan morphogenesis. Additionally, we identified six other factors that target the histone gene array: JIL-1, hormone-like receptor 78 (Hr78), the long isoform of female sterile homeotic (1) (fs(1)h) as well as the general transcription factors TBP associated factor 1 (TAF-1), Transcription Factor IIB (TFIIB), and Transcription Factor IIF (TFIIF).
Conclusions: Our foundational screen provides several candidates for future studies into factors that may influence histone biogenesis. Further, our study emphasizes the powerful reservoir of publicly available datasets, which can be mined as a primary screening technique.
Keywords: ChIP-seq; Course-based Undergraduate Research Experience; Drosophila; Galaxy; Histone locus; Histone locus body; Hox factors.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
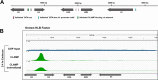
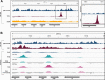
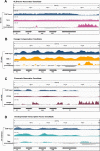
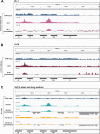

Update of
-
A bioinformatics screen reveals Hox and chromatin remodeling factors at the Drosophila histone locus.bioRxiv [Preprint]. 2023 Jan 6:2023.01.06.523008. doi: 10.1101/2023.01.06.523008. bioRxiv. 2023. Update in: BMC Genom Data. 2023 Sep 21;24(1):54. doi: 10.1186/s12863-023-01147-0. PMID: 36711759 Free PMC article. Updated. Preprint.
Similar articles
-
A bioinformatics screen reveals Hox and chromatin remodeling factors at the Drosophila histone locus.bioRxiv [Preprint]. 2023 Jan 6:2023.01.06.523008. doi: 10.1101/2023.01.06.523008. bioRxiv. 2023. Update in: BMC Genom Data. 2023 Sep 21;24(1):54. doi: 10.1186/s12863-023-01147-0. PMID: 36711759 Free PMC article. Updated. Preprint.
-
Hox gene Ultrabithorax regulates distinct sets of target genes at successive stages of Drosophila haltere morphogenesis.Proc Natl Acad Sci U S A. 2011 Feb 15;108(7):2855-60. doi: 10.1073/pnas.1015077108. Epub 2011 Jan 31. Proc Natl Acad Sci U S A. 2011. PMID: 21282633 Free PMC article.
-
Genome-wide tissue-specific occupancy of the Hox protein Ultrabithorax and Hox cofactor Homothorax in Drosophila.PLoS One. 2011 Apr 5;6(4):e14686. doi: 10.1371/journal.pone.0014686. PLoS One. 2011. PMID: 21483663 Free PMC article.
-
Cellular analysis of newly identified Hox downstream genes in Drosophila.Eur J Cell Biol. 2010 Feb-Mar;89(2-3):273-8. doi: 10.1016/j.ejcb.2009.11.012. Epub 2009 Dec 16. Eur J Cell Biol. 2010. PMID: 20018403 Review.
-
The bithorax complex of Drosophila an exceptional Hox cluster.Curr Top Dev Biol. 2009;88:1-33. doi: 10.1016/S0070-2153(09)88001-0. Curr Top Dev Biol. 2009. PMID: 19651300 Review.
Cited by
-
Histone locus bodies: a paradigm for how nuclear biomolecular condensates control cell cycle regulated gene expression.Nucleus. 2023 Dec;14(1):2293604. doi: 10.1080/19491034.2023.2293604. Epub 2023 Dec 14. Nucleus. 2023. PMID: 38095604 Free PMC article. Review.
-
Cis element length variability does not confer differential transcription factor occupancy at the D. melanogaster histone locus.bioRxiv [Preprint]. 2024 Jun 28:2024.06.24.600460. doi: 10.1101/2024.06.24.600460. bioRxiv. 2024. PMID: 38979213 Free PMC article. Preprint.
-
Sequence reliance of a Drosophila context-dependent transcription factor.bioRxiv [Preprint]. 2023 Dec 8:2023.12.07.570650. doi: 10.1101/2023.12.07.570650. bioRxiv. 2023. Update in: Genetics. 2024 Jul 8;227(3):iyae060. doi: 10.1093/genetics/iyae060. PMID: 38106168 Free PMC article. Updated. Preprint.
-
Zelda is dispensable for Drosophila melanogaster histone gene regulation.Mol Biol Cell. 2025 Feb 1;36(2):br3. doi: 10.1091/mbc.E24-01-0028. Epub 2024 Dec 11. Mol Biol Cell. 2025. PMID: 39661467 Free PMC article.
-
Cell cycle-regulated transcriptional pausing of Drosophila replication-dependent histone genes.bioRxiv [Preprint]. 2024 Dec 17:2024.12.16.628706. doi: 10.1101/2024.12.16.628706. bioRxiv. 2024. Update in: Mol Biol Cell. 2025 Jul 1;36(7):ar88. doi: 10.1091/mbc.E25-05-0212. PMID: 39763942 Free PMC article. Updated. Preprint.
References
-
- Arias Escayola D, Neugebauer KM. Dynamics and function of Nuclear Bodies during Embryogenesis. Biochemistry. 2018;57(17):2462–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
