U2AF1 pathogenic variants in myeloid neoplasms and precursor states: distribution of co-mutations and prognostic heterogeneity
- PMID: 37735430
- PMCID: PMC10514309
- DOI: 10.1038/s41408-023-00922-7
U2AF1 pathogenic variants in myeloid neoplasms and precursor states: distribution of co-mutations and prognostic heterogeneity
Abstract
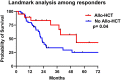
We have previously recognized the genotypic and prognostic heterogeneity of U2AF1 mutations (MT) in myelofibrosis (MF) and myelodysplastic syndromes (MDS). In the current study, we considered 179 U2AF1-mutated patients with clonal cytopenia of undetermined significance (CCUS; n = 22), MDS (n = 108), MDS/acute myeloid leukemia (AML; n = 18) and AML (n = 31). U2AF1 variants included S34 (60%), Q157 (35%), and others (5%): corresponding mutational frequencies were 45%, 55%, and 0% in CCUS; 57%, 39%, and 4% in MDS; 61%, 33%, and 6% in MDS/AML; and 55%, 35% and 10% in AML (P = 0.17, 0.36 and 0.09), respectively. Concurrent mutations included ASXL1 (37%), BCOR (19%), RUNX1 (14%), TET2 (15%), DNMT3A (10%), NRAS/KRAS (8%), TP53 (8%), JAK2 (5.5%) and SETBP1 (5%). The two most frequent U2AF1 MT were S34F (n = 97) and Q157P (n = 46); concurrent MT were more likely to be seen with the latter (91% vs 74%; P = 0.01) and abnormal karyotype with the former (70% vs 62%; P = 0.05). U2AF1 S34F MT clustered with BCOR (P = 0.04) and Q157P MT with ASXL1 (P = 0.01) and TP53 (P = 0.03). The median overall survival (OS) in months was significantly worse in AML (14.2) vs MDS/AML (27.3) vs MDS (33.7; P = 0.001); the latter had similar OS with CCUS (30.0). In morphologically high-risk disease (n = 49), defined by ≥10% blood or bone marrow blasts (i.e., AML or MDS/AML), median OS was 14.2 with Q157P vs 37.1 months in the presence of S34F (P = 0.008); transplant-adjusted multivariable analysis confirmed the detrimental impact of Q157P (P = 0.01) on survival and also identified JAK2 MT as an additional risk factor (P = 0.02). OS was favorably affected by allogeneic hematopoietic stem cell transplantation (HR: 0.16, 95% CI; 0.04-0.61, P = 0.007). The current study defines the prevalence and co-mutational profiles of U2AF1 pathogenic variants in AML, MDS/AML, MDS, and CCUS, and suggests prognostic heterogeneity in patients with ≥10% blasts.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Reinig E, Yang F, Traer E, Arora R, Brown S, Rattray R, et al. Targeted next-generation sequencing in myelodysplastic syndrome and chronic myelomonocytic leukemia aids diagnosis in challenging cases and identifies frequent spliceosome mutations in transformed acute myeloid leukemia. Am J Clin Pathol. 2016;145:497–506. doi: 10.1093/ajcp/aqw016. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

