Nematostella vectensis exemplifies the exceptional expansion and diversity of opsins in the eyeless Hexacorallia
- PMID: 37735470
- PMCID: PMC10512536
- DOI: 10.1186/s13227-023-00218-8
Nematostella vectensis exemplifies the exceptional expansion and diversity of opsins in the eyeless Hexacorallia
Abstract
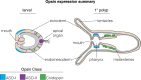
Background: Opsins are the primary proteins responsible for light detection in animals. Cnidarians (jellyfish, sea anemones, corals) have diverse visual systems that have evolved in parallel with bilaterians (squid, flies, fish) for hundreds of millions of years. Medusozoans (e.g., jellyfish, hydroids) have evolved eyes multiple times, each time independently incorporating distinct opsin orthologs. Anthozoans (e.g., corals, sea anemones,) have diverse light-mediated behaviors and, despite being eyeless, exhibit more extensive opsin duplications than medusozoans. To better understand the evolution of photosensitivity in animals without eyes, we increased anthozoan representation in the phylogeny of animal opsins and investigated the large but poorly characterized opsin family in the sea anemone Nematostella vectensis.
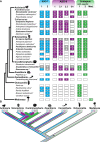
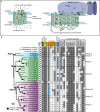
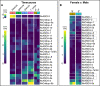
Results: We analyzed genomic and transcriptomic data from 16 species of cnidarians to generate a large opsin phylogeny (708 sequences) with the largest sampling of anthozoan sequences to date. We identified 29 opsins from N. vectensis (NvOpsins) with high confidence, using transcriptomic and genomic datasets. We found that lineage-specific opsin duplications are common across Cnidaria, with anthozoan lineages exhibiting among the highest numbers of opsins in animals. To establish putative photosensory function of NvOpsins, we identified canonically conserved protein domains and amino acid sequences essential for opsin function in other animal species. We show high sequence diversity among NvOpsins at sites important for photoreception and transduction, suggesting potentially diverse functions. We further examined the spatiotemporal expression of NvOpsins and found both dynamic expression of opsins during embryonic development and sexually dimorphic opsin expression in adults.
Conclusions: These data show that lineage-specific duplication and divergence has led to expansive diversity of opsins in eyeless cnidarians, suggesting opsins from these animals may exhibit novel biochemical functions. The variable expression patterns of opsins in N. vectensis suggest opsin gene duplications allowed for a radiation of unique sensory cell types with tissue- and stage-specific functions. This diffuse network of distinct sensory cell types could be an adaptive solution for varied sensory tasks experienced in distinct life history stages in Anthozoans.
Keywords: Anthozoa; Cnidaria; Hexacorallia; Nematostella; Opsin; Photoreceptor; Rhodopsin; Sea anemone.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
Larval swimming in the sea anemone Nematostella vectensis is sensitive to a broad light spectrum and exhibits a wavelength-dependent behavioral switch.Ecol Evol. 2024 Apr 15;14(4):e11222. doi: 10.1002/ece3.11222. eCollection 2024 Apr. Ecol Evol. 2024. PMID: 38628921 Free PMC article.
-
De novo genome assembly of the Edwardsiid anthozoan Edwardsia elegans.G3 (Bethesda). 2025 Apr 17;15(4):jkaf011. doi: 10.1093/g3journal/jkaf011. G3 (Bethesda). 2025. PMID: 39849905 Free PMC article.
-
Diurnal and circadian regulation of opsin-like transcripts in the eyeless cnidarian Hydra.Biomol Concepts. 2024 Mar 19;15(1). doi: 10.1515/bmc-2022-0044. eCollection 2024 Jan 1. Biomol Concepts. 2024. PMID: 38502542
-
The diversity of invertebrate visual opsins spanning Protostomia, Deuterostomia, and Cnidaria.Dev Biol. 2022 Dec;492:187-199. doi: 10.1016/j.ydbio.2022.10.011. Epub 2022 Oct 19. Dev Biol. 2022. PMID: 36272560 Free PMC article. Review.
-
Shedding new light on opsin evolution.Proc Biol Sci. 2012 Jan 7;279(1726):3-14. doi: 10.1098/rspb.2011.1819. Epub 2011 Oct 19. Proc Biol Sci. 2012. PMID: 22012981 Free PMC article. Review.
Cited by
-
Larval swimming in the sea anemone Nematostella vectensis is sensitive to a broad light spectrum and exhibits a wavelength-dependent behavioral switch.Ecol Evol. 2024 Apr 15;14(4):e11222. doi: 10.1002/ece3.11222. eCollection 2024 Apr. Ecol Evol. 2024. PMID: 38628921 Free PMC article.
-
Sea Anemone Membrane Attack Complex/Perforin Superfamily Demonstrates an Evolutionary Transitional State between Venomous and Developmental Functions.Mol Biol Evol. 2024 May 3;41(5):msae082. doi: 10.1093/molbev/msae082. Mol Biol Evol. 2024. PMID: 38676945 Free PMC article.
-
Genome assembly of Bougainvillia cf. muscus (Cnidaria: Hydrozoa).G3 (Bethesda). 2025 Jul 9;15(7):jkaf110. doi: 10.1093/g3journal/jkaf110. G3 (Bethesda). 2025. PMID: 40388381 Free PMC article.
-
CLOCK evolved in cnidaria to synchronize internal rhythms with diel environmental cues.Elife. 2024 May 14;12:RP89499. doi: 10.7554/eLife.89499. Elife. 2024. PMID: 38743049 Free PMC article.
-
Phototactic preference and its genetic basis in the planulae of the colonial Hydrozoan Hydractinia symbiolongicarpus.bioRxiv [Preprint]. 2024 Apr 1:2024.03.28.585045. doi: 10.1101/2024.03.28.585045. bioRxiv. 2024. PMID: 38617216 Free PMC article. Preprint.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

