The NCOR-HDAC3 co-repressive complex modulates the leukemogenic potential of the transcription factor ERG
- PMID: 37735473
- PMCID: PMC10514085
- DOI: 10.1038/s41467-023-41067-2
The NCOR-HDAC3 co-repressive complex modulates the leukemogenic potential of the transcription factor ERG
Abstract
The ERG (ETS-related gene) transcription factor is linked to various types of cancer, including leukemia. However, the specific ERG domains and co-factors contributing to leukemogenesis are poorly understood. Drug targeting a transcription factor such as ERG is challenging. Our study reveals the critical role of a conserved amino acid, proline, at position 199, located at the 3' end of the PNT (pointed) domain, in ERG's ability to induce leukemia. P199 is necessary for ERG to promote self-renewal, prevent myeloid differentiation in hematopoietic progenitor cells, and initiate leukemia in mouse models. Here we show that P199 facilitates ERG's interaction with the NCoR-HDAC3 co-repressor complex. Inhibiting HDAC3 reduces the growth of ERG-dependent leukemic and prostate cancer cells, indicating that the interaction between ERG and the NCoR-HDAC3 co-repressor complex is crucial for its oncogenic activity. Thus, targeting this interaction may offer a potential therapeutic intervention.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures


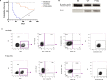

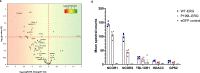




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

