Characterization of tumor microenvironment and tumor immunology based on the double-stranded RNA-binding protein related genes in cervical cancer
- PMID: 37735483
- PMCID: PMC10515034
- DOI: 10.1186/s12967-023-04505-9
Characterization of tumor microenvironment and tumor immunology based on the double-stranded RNA-binding protein related genes in cervical cancer
Abstract
Background: Cervical cancer is one of the most common gynecological cancers threatening women's health worldwide. Double-stranded RNA-binding proteins (dsRBPs) regulate innate immunity and are therefore believed to be involved in virus-related malignancies, however, their role in cervical cancer is not well known.
Methods: We performed RNA-seq of tumor samples from cervical cancer patients in local cohort and also assessed the RNA-seq and clinical data derived from public datasets. By using single sample Gene Set Enrichment Analysis (ssGSEA) and univariate Cox analysis, patients were stratified into distinct dsRBP clusters. Stepwise Cox and CoxBoost were performed to construct a risk model based on optimal dsRBPs clusters-related differentially expressed genes (DEGs), and GSE44001 and CGCI-HTMCP-CC were employed as two external validation cohorts. Single cell RNA sequencing data from GSE168652 and Scissor algorithm were applied to evaluated the signature-related cell population.
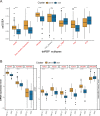
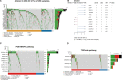
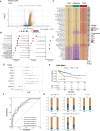
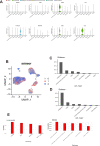
Results: The expression of dsRBP features was found to be associated with HPV infection and carcinogenesis in CESC. However, only Adenosine deaminases acting on RNA (ADAR) and Dicer, Drosha, and Argonautes (DDR) exhibited significant correlations with the overall survival (OS) of CESC patients. Based on these findings, CESC patients were divided into three dsRBP clusters. Cluster 3 showed superior OS but lower levels of ADAR and DDR. Additionally, Cluster 3 demonstrated enhanced innate immunity, with significantly higher activity in cancer immunity cycles, immune scores, and levels of tumor-infiltrating immune cells, particularly CD8+ T cells. Furthermore, a risk model based on nine dsRBP cluster-related DEGs was established. The accuracy of survival prediction for 1 to 5 years was consistently above 0.78, and this model's robust predictive capacity was confirmed by two external validation sets. The low-risk group exhibited significantly higher levels of immune checkpoints, such as PDCD1 and CTLA4, as well as a higher abundance of CD8+ T cells. Analysis of single-cell sequencing data revealed a significant association between the dsRBP signature and glycolysis. Importantly, low-risk patients showed improved OS and a higher response rate to immunotherapy, along with enduring clinical benefits from concurrent chemoradiotherapy.
Conclusions: dsRBP played a crucial role in the regulation of prognosis and tumor immunology in cervical cancer, and its prognostic signature provides a strategy for risk stratification and immunotherapy evaluation.
Keywords: Cervical cancer; Chemotherapy; Immunotherapy; Risk model; Tumor microenvironment; dsRBP.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures







Similar articles
-
A Novel Prognostic Risk Model for Cervical Cancer Based on Immune Checkpoint HLA-G-Driven Differentially Expressed Genes.Front Immunol. 2022 Jul 18;13:851622. doi: 10.3389/fimmu.2022.851622. eCollection 2022. Front Immunol. 2022. PMID: 35924232 Free PMC article.
-
A novel DNA damage repair-related gene signature predicting survival, immune infiltration and drug sensitivity in cervical cancer based on single cell sequencing.Front Immunol. 2023 Jun 28;14:1198391. doi: 10.3389/fimmu.2023.1198391. eCollection 2023. Front Immunol. 2023. PMID: 37449209 Free PMC article.
-
A comprehensive study of oxidative stress-related effects on the prognosis and drug therapy of cervical cancer.J Gene Med. 2024 Jan;26(1):e3581. doi: 10.1002/jgm.3581. Epub 2023 Aug 22. J Gene Med. 2024. PMID: 37605936
-
Identification of prognostic biomarkers for cervical cancer based on programmed cell death-related genes and assessment of their immune profile and response to drug therapy.J Gene Med. 2024 Jan;26(1):e3643. doi: 10.1002/jgm.3643. Epub 2023 Dec 4. J Gene Med. 2024. PMID: 38044747
-
Infiltration Patterns of Cervical Epithelial Microenvironment Cells During Carcinogenesis.Front Immunol. 2022 Jul 14;13:888176. doi: 10.3389/fimmu.2022.888176. eCollection 2022. Front Immunol. 2022. PMID: 35911729 Free PMC article. Review.
Cited by
-
Ferroptosis-related gene transferrin receptor protein 1 expression correlates with the prognosis and tumor immune microenvironment in cervical cancer.PeerJ. 2024 Aug 6;12:e17842. doi: 10.7717/peerj.17842. eCollection 2024. PeerJ. 2024. PMID: 39131609 Free PMC article.
-
The benefit and risk of addition of PD-1/PD-L1 inhibitors to chemotherapy for advanced cervical cancer: a phase 3 randomized controlled trials based meta-analysis.BMC Cancer. 2025 Mar 12;25(1):450. doi: 10.1186/s12885-025-13843-4. BMC Cancer. 2025. PMID: 40075324 Free PMC article.
References
-
- Melnikow J, Henderson JT, Burda BU, Senger CA, Durbin S, Weyrich MS. Screening for cervical cancer with high-risk human papillomavirus testing: updated evidence report and systematic review for the US Preventive Services Task Force. JAMA. 2018;320(7):687–705. doi: 10.1001/jama.2018.10400. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials

