Characterization and organelle genome sequencing of Pyropia species from Myanmar
- PMID: 37735516
- PMCID: PMC10514050
- DOI: 10.1038/s41598-023-42262-3
Characterization and organelle genome sequencing of Pyropia species from Myanmar
Abstract
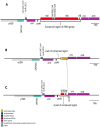
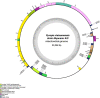
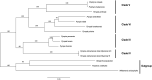
Pyropia is a genus comprising red algae of the Bangiaceae family that is commonly found in intertidal zones worldwide. However, understanding of Pyropia species that are prone to tropical regions remains limited despite recent breakthroughs in genomic research. Within the realm of Pyropia species thriving in tropical regions, P. vietnamensis stands out as a widely recognized species. In this study, we aimed to investigate Pyropia species in the southwest coast of Myanmar using physiological and molecular approaches, culture-based analyses, chloroplast rbcL and nuclear SSU gene sequencing, and whole chloroplast and mitochondrial genome sequencing. Physiological analysis showed that the Myanmar samples were more heat-tolerant than their Japanese counterparts, including those of subtropical origin. Additionally, molecular characterization revealed that the Myanmar samples were closely related to P. vietnamensis from India. This study is the first to sequence the chloroplast and mitochondrial genomes of Pyropia species from tropical regions. A unique deletion event was observed within a ribosomal RNA gene cluster in the chloroplast genome of the studied Pyropia species, which is a deviation from the usual characteristics of most Pyropia species. This study improves current understanding of the physiological and molecular characteristics of this comparatively understudied Pyropia species that grows in tropical regions.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Yang LE, et al. Redefining Pyropia (Bangiales, Rhodophyta): Four new genera, resurrection of Porphyrella and description of Calidia pseudolobata sp. Nov. from China. J. Phycol. 2020;56:862–879. - PubMed
-
- Mumford Jr, T. F. Porphyra as Food: Cultivation and Economics in Algae and Human Affairs (ed. Lembi, C. A. & Waaland, J. R.) 87–117 (Cambridge Univ., 1988).
-
- FAO Fisheries and Aquaculture Information and Statistics Service. FAO Yearbook: Fishery and Agriculture Statistics 2019 (United Nations Publs, 2021).
-
- FAO (Food and Agriculture Organization of the United Nations). The state of world fisheries and aquaculture. http://www.fao.org/3/ca9229en/CA9229EN.pdf, (2020).
-
- Cian RE, Martínez-Augustin O, Drago SR. Bioactive properties of peptides obtained by enzymatic hydrolysis from protein byproducts of Porphyra columbina. Food Res. Int. 2012;49:364–372.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

