Regulation of cellular cholesterol distribution via non-vesicular lipid transport at ER-Golgi contact sites
- PMID: 37735529
- PMCID: PMC10514280
- DOI: 10.1038/s41467-023-41213-w
Regulation of cellular cholesterol distribution via non-vesicular lipid transport at ER-Golgi contact sites
Abstract
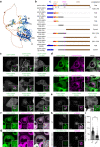
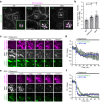
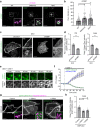
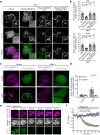
Abnormal distribution of cellular cholesterol is associated with numerous diseases, including cardiovascular and neurodegenerative diseases. Regulated transport of cholesterol is critical for maintaining its proper distribution in the cell, yet the underlying mechanisms remain unclear. Here, we show that lipid transfer proteins, namely ORP9, OSBP, and GRAMD1s/Asters (GRAMD1a/GRAMD1b/GRAMD1c), control non-vesicular cholesterol transport at points of contact between the ER and the trans-Golgi network (TGN), thereby maintaining cellular cholesterol distribution. ORP9 localizes to the TGN via interaction between its tandem α-helices and ORP10/ORP11. ORP9 extracts PI4P from the TGN to prevent its overaccumulation and suppresses OSBP-mediated PI4P-driven cholesterol transport to the Golgi. By contrast, GRAMD1s transport excess cholesterol from the Golgi to the ER, thereby preventing its build-up. Cells lacking ORP9 exhibit accumulation of cholesterol at the Golgi, which is further enhanced by additional depletion of GRAMD1s with major accumulation in the plasma membrane. This is accompanied by chronic activation of the SREBP-2 signalling pathway. Our findings reveal the importance of regulated lipid transport at ER-Golgi contacts for maintaining cellular cholesterol distribution and homeostasis.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
ORP9 and ORP10 form a heterocomplex to transfer phosphatidylinositol 4-phosphate at ER-TGN contact sites.Cell Mol Life Sci. 2023 Feb 28;80(3):77. doi: 10.1007/s00018-023-04728-5. Cell Mol Life Sci. 2023. PMID: 36853333 Free PMC article.
-
Oxysterol binding protein-related Protein 9 (ORP9) is a cholesterol transfer protein that regulates Golgi structure and function.Mol Biol Cell. 2009 Mar;20(5):1388-99. doi: 10.1091/mbc.e08-09-0905. Epub 2009 Jan 7. Mol Biol Cell. 2009. PMID: 19129476 Free PMC article.
-
Sterol transfer, PI4P consumption, and control of membrane lipid order by endogenous OSBP.EMBO J. 2017 Nov 2;36(21):3156-3174. doi: 10.15252/embj.201796687. Epub 2017 Oct 4. EMBO J. 2017. PMID: 28978670 Free PMC article.
-
GRAMD1-mediated accessible cholesterol sensing and transport.Biochim Biophys Acta Mol Cell Biol Lipids. 2021 Aug;1866(8):158957. doi: 10.1016/j.bbalip.2021.158957. Epub 2021 Apr 28. Biochim Biophys Acta Mol Cell Biol Lipids. 2021. PMID: 33932585 Review.
-
Lipid exchange and signaling at ER-Golgi contact sites.Curr Opin Cell Biol. 2019 Apr;57:8-15. doi: 10.1016/j.ceb.2018.10.002. Epub 2018 Nov 1. Curr Opin Cell Biol. 2019. PMID: 30390465 Review.
Cited by
-
Getting to Grips with the Oxysterol-Binding Protein Family - a Forty Year Perspective.Contact (Thousand Oaks). 2024 Aug 28;7:25152564241273598. doi: 10.1177/25152564241273598. eCollection 2024 Jan-Dec. Contact (Thousand Oaks). 2024. PMID: 39210909 Free PMC article. Review.
-
Salmonella Typhimurium effector SseI regulates host peroxisomal dynamics to acquire lysosomal cholesterol.EMBO Rep. 2025 Feb;26(3):656-689. doi: 10.1038/s44319-024-00328-x. Epub 2024 Dec 18. EMBO Rep. 2025. PMID: 39695325 Free PMC article.
-
OSBP is a major determinant of Golgi phosphatidylinositol 4-phosphate homeostasis.bioRxiv [Preprint]. 2024 Jan 25:2023.12.21.572879. doi: 10.1101/2023.12.21.572879. bioRxiv. 2024. Update in: Contact (Thousand Oaks). 2024 Feb 22;7:25152564241232196. doi: 10.1177/25152564241232196. PMID: 38187665 Free PMC article. Updated. Preprint.
-
OSBP is a Major Determinant of Golgi Phosphatidylinositol 4-Phosphate Homeostasis.Contact (Thousand Oaks). 2024 Feb 22;7:25152564241232196. doi: 10.1177/25152564241232196. eCollection 2024 Jan-Dec. Contact (Thousand Oaks). 2024. PMID: 38405037 Free PMC article.
-
Visualization of accessible cholesterol using a GRAM domain-based biosensor.Nat Commun. 2023 Oct 25;14(1):6773. doi: 10.1038/s41467-023-42498-7. Nat Commun. 2023. PMID: 37880244 Free PMC article.
References
-
- Vance JE. Phospholipid synthesis and transport in mammalian cells. Traffic. 2015;16:1–18. - PubMed
-
- Norman AW, Mizwicki MT, Norman DP. Steroid-hormone rapid actions, membrane receptors and a conformational ensemble model. Nat. Rev. Drug Discov. 2004;3:27–41. - PubMed
-
- Dai L, et al. Cholesterol metabolism in neurodegenerative diseases: molecular mechanisms and therapeutic targets. Mol. Neurobiol. 2021;58:2183–2201. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

