An experimental comparison between primer and nucleotide labelling to produce RPA-amplicons used for multiplex detection of antibiotic resistance genes
- PMID: 37735542
- PMCID: PMC10514322
- DOI: 10.1038/s41598-023-42830-7
An experimental comparison between primer and nucleotide labelling to produce RPA-amplicons used for multiplex detection of antibiotic resistance genes
Abstract
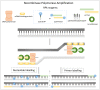
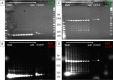
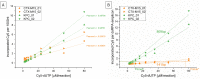
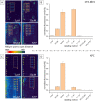
Direct labelling of amplification products using isothermal amplification is currently done most frequently by incorporating previously labelled primer. Although this method is well proven and widely used, it is not a universal solution due to some weaknesses. Alternatively, labelled nucleotides could be used, whose application and functionality have been already partially demonstrated. It remains to be determined how this method performs in comparison to traditional labelling, in particular combined with isothermal amplification methods. In this work, we show a detailed analysis of the labelling efficiency under different conditions and compare the results with the traditional primer-labelling method in the context of RPA amplification. Impressively, our results showed that using Cy5-labelled dUTPs can achieve much more efficient labelling for fragments above 200 bp, while using them for smaller fragments does not bring any relevant disadvantages, but also no major benefit. Furthermore, this work successfully demonstrate for the first time a quadruplex microarray for the detection of resistance genes using RPA and direct labelling with Cy5-dUTP as a potential application scenario. The sensitivities achieved here extend to SNP discovery for the detection of the proper blaKPC variant.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Comparison of pre-labelled primers and nucleotides as DNA labelling method for lateral flow detection of Legionella pneumophila amplicons.Sci Rep. 2024 Feb 29;14(1):5018. doi: 10.1038/s41598-024-55703-4. Sci Rep. 2024. PMID: 38424185 Free PMC article.
-
Using Cy5-dUTP labelling of RPA-amplicons with downstream microarray analysis for the detection of antibiotic resistance genes.Sci Rep. 2021 Oct 11;11(1):20137. doi: 10.1038/s41598-021-99774-z. Sci Rep. 2021. PMID: 34635776 Free PMC article.
-
Investigation and validation of labelling loop mediated isothermal amplification (LAMP) products with different nucleotide modifications for various downstream analysis.Sci Rep. 2022 May 3;12(1):7137. doi: 10.1038/s41598-022-11320-7. Sci Rep. 2022. PMID: 35504953 Free PMC article.
-
Recent advances in recombinase polymerase amplification: Principle, advantages, disadvantages and applications.Front Cell Infect Microbiol. 2022 Nov 28;12:1019071. doi: 10.3389/fcimb.2022.1019071. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36519130 Free PMC article. Review.
-
The single-nucleotide primer extension (SNuPE) method for the multiplex detection of various DNA sequences: from detection of point mutations to microbial ecology.Biochem Soc Trans. 2009 Apr;37(Pt 2):454-9. doi: 10.1042/BST0370454. Biochem Soc Trans. 2009. PMID: 19290881 Review.
Cited by
-
Comparison of pre-labelled primers and nucleotides as DNA labelling method for lateral flow detection of Legionella pneumophila amplicons.Sci Rep. 2024 Feb 29;14(1):5018. doi: 10.1038/s41598-024-55703-4. Sci Rep. 2024. PMID: 38424185 Free PMC article.
-
Direct TAMRA-dUTP labeling of M. tuberculosis genes using loop-mediated isothermal amplification (LAMP).Sci Rep. 2024 Mar 7;14(1):5611. doi: 10.1038/s41598-024-55289-x. Sci Rep. 2024. PMID: 38454089 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

