Multimodal single-cell datasets characterize antigen-specific CD8+ T cells across SARS-CoV-2 vaccination and infection
- PMID: 37735591
- PMCID: PMC10522491
- DOI: 10.1038/s41590-023-01608-9
Multimodal single-cell datasets characterize antigen-specific CD8+ T cells across SARS-CoV-2 vaccination and infection
Abstract
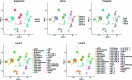
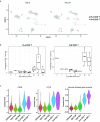
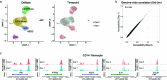
The immune response to SARS-CoV-2 antigen after infection or vaccination is defined by the durable production of antibodies and T cells. Population-based monitoring typically focuses on antibody titer, but there is a need for improved characterization and quantification of T cell responses. Here, we used multimodal sequencing technologies to perform a longitudinal analysis of circulating human leukocytes collected before and after immunization with the mRNA vaccine BNT162b2. Our data indicated distinct subpopulations of CD8+ T cells, which reliably appeared 28 days after prime vaccination. Using a suite of cross-modality integration tools, we defined their transcriptome, accessible chromatin landscape and immunophenotype, and we identified unique biomarkers within each modality. We further showed that this vaccine-induced population was SARS-CoV-2 antigen-specific and capable of rapid clonal expansion. Moreover, we identified these CD8+ T cell populations in scRNA-seq datasets from COVID-19 patients and found that their relative frequency and differentiation outcomes were predictive of subsequent clinical outcomes.
© 2023. The Author(s).
Conflict of interest statement
In the past 3 years, R.S. has worked as a consultant for Bristol-Myers Squibb, Regeneron and Kallyope and served as a scientific advisory board member for ImmunAI, Resolve Biosciences, Nanostring and the NYC Pandemic Response Lab. R.S. and Y.H. are co-founders and equity holders of Neptune Bio. As of August 1, 2023, Y.H. is an employee of Neptune Bio. D.R.L. is cofounder of Vedanta Biosciences and ImmunAI, on the advisory boards of IMIDomics and Evommune and on the board of directors of Pfizer. M.J.M. reports potential competing interests: laboratory research and clinical trials contracts with Lilly, Pfizer (exclusive of the current work) and Sanofi for vaccines or MAB vs SARS-CoV-2; contract funding from USG/HHS/BARDA for research specimen characterization and repository; research grant funding from USG/HHS/NIH for SARS-CoV-2 vaccine and MAB clinical trials; and personal fees from Meissa Vaccines and Pfizer for scientific advisory board service. R.S.H. has received research support from CareDx for SARS-CoV-2 vaccine studies. R.S.H. is a consultant for Bristol-Myers-Squibb. All other authors declare no competing interests.
Figures






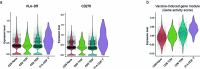


Update of
-
Multimodal characterization of antigen-specific CD8 + T cells across SARS-CoV-2 vaccination and infection.bioRxiv [Preprint]. 2023 Jan 24:2023.01.24.525203. doi: 10.1101/2023.01.24.525203. bioRxiv. 2023. Update in: Nat Immunol. 2023 Oct;24(10):1725-1734. doi: 10.1038/s41590-023-01608-9. PMID: 36747786 Free PMC article. Updated. Preprint.
Comment in
-
Vaccine-induced CD8+ T cells are key to protection from SARS-CoV-2.Nat Immunol. 2023 Oct;24(10):1594-1596. doi: 10.1038/s41590-023-01621-y. Nat Immunol. 2023. PMID: 37735590 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

