Dispersal, habitat filtering, and eco-evolutionary dynamics as drivers of local and global wetland viral biogeography
- PMID: 37735616
- PMCID: PMC10579374
- DOI: 10.1038/s41396-023-01516-8
Dispersal, habitat filtering, and eco-evolutionary dynamics as drivers of local and global wetland viral biogeography
Abstract
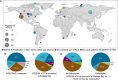
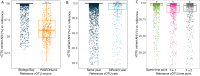
Wetlands store 20-30% of the world's soil carbon, and identifying the microbial controls on these carbon reserves is essential to predicting feedbacks to climate change. Although viral infections likely play important roles in wetland ecosystem dynamics, we lack a basic understanding of wetland viral ecology. Here 63 viral size-fraction metagenomes (viromes) and paired total metagenomes were generated from three time points in 2021 at seven fresh- and saltwater wetlands in the California Bodega Marine Reserve. We recovered 12,826 viral population genomic sequences (vOTUs), only 4.4% of which were detected at the same field site two years prior, indicating a small degree of population stability or recurrence. Viral communities differed most significantly among the seven wetland sites and were also structured by habitat (plant community composition and salinity). Read mapping to a new version of our reference database, PIGEONv2.0 (515,763 vOTUs), revealed 196 vOTUs present over large geographic distances, often reflecting shared habitat characteristics. Wetland vOTU microdiversity was significantly lower locally than globally and lower within than between time points, indicating greater divergence with increasing spatiotemporal distance. Viruses tended to have broad predicted host ranges via CRISPR spacer linkages to metagenome-assembled genomes, and increased SNP frequencies in CRISPR-targeted major tail protein genes suggest potential viral eco-evolutionary dynamics in response to both immune targeting and changes in host cell receptors involved in viral attachment. Together, these results highlight the importance of dispersal, environmental selection, and eco-evolutionary dynamics as drivers of local and global wetland viral biogeography.
© 2023. The Author(s), under exclusive licence to International Society for Microbial Ecology.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Minnesota peat viromes reveal terrestrial and aquatic niche partitioning for local and global viral populations.Microbiome. 2021 Nov 26;9(1):233. doi: 10.1186/s40168-021-01156-0. Microbiome. 2021. PMID: 34836550 Free PMC article.
-
Soil Viruses Are Underexplored Players in Ecosystem Carbon Processing.mSystems. 2018 Oct 2;3(5):e00076-18. doi: 10.1128/mSystems.00076-18. eCollection 2018 Sep-Oct. mSystems. 2018. PMID: 30320215 Free PMC article.
-
Climate and plant controls on soil organic matter in coastal wetlands.Glob Chang Biol. 2018 Nov;24(11):5361-5379. doi: 10.1111/gcb.14376. Epub 2018 Jul 29. Glob Chang Biol. 2018. PMID: 29957880
-
Impacts of human-induced environmental change in wetlands on aquatic animals.Biol Rev Camb Philos Soc. 2018 Feb;93(1):529-554. doi: 10.1111/brv.12358. Epub 2017 Sep 19. Biol Rev Camb Philos Soc. 2018. PMID: 28929570 Review.
-
A review of factors affecting the soil microbial community structure in wetlands.Environ Sci Pollut Res Int. 2024 Jul;31(34):46760-46768. doi: 10.1007/s11356-024-34132-w. Epub 2024 Jul 5. Environ Sci Pollut Res Int. 2024. PMID: 38967845 Review.
Cited by
-
Patchy burn severity explains heterogeneous soil viral and prokaryotic responses to fire in a mixed conifer forest.mSystems. 2025 Jun 17;10(6):e0174924. doi: 10.1128/msystems.01749-24. Epub 2025 May 14. mSystems. 2025. PMID: 40366158 Free PMC article.
-
Soil viral communities shifted significantly after wildfire in chaparral and woodland habitats.ISME Commun. 2025 May 6;5(1):ycaf073. doi: 10.1093/ismeco/ycaf073. eCollection 2025 Jan. ISME Commun. 2025. PMID: 40385308 Free PMC article.
-
Community Structure, Drivers, and Potential Functions of Different Lifestyle Viruses in Chaohu Lake.Viruses. 2024 Apr 11;16(4):590. doi: 10.3390/v16040590. Viruses. 2024. PMID: 38675931 Free PMC article.
-
Targeted hypermutation of putative antigen sensors in multicellular bacteria.Proc Natl Acad Sci U S A. 2024 Feb 27;121(9):e2316469121. doi: 10.1073/pnas.2316469121. Epub 2024 Feb 14. Proc Natl Acad Sci U S A. 2024. PMID: 38354254 Free PMC article.
-
Targeted viromes and total metagenomes capture distinct components of bee gut phage communities.Microbiome. 2024 Aug 23;12(1):155. doi: 10.1186/s40168-024-01875-0. Microbiome. 2024. PMID: 39175056 Free PMC article.
References
-
- Hopkinson CS, Cai W-J, Hu X. Carbon sequestration in wetland dominated coastal systems—a global sink of rapidly diminishing magnitude. Curr Opin Environ Sustainability. 2012;4:186–94.
-
- Mitsch WJ, Gosselink JG. The value of wetlands: importance of scale and landscape setting. Ecol Econ. 2000;35:25–33.
-
- Mitsch WJ, Bernal B, Hernandez ME. Ecosystem services of wetlands. Int J Biodivers Sci Eco Services Mgmt. 2015;11:1–4.
-
- Yarwood SA. The role of wetland microorganisms in plant-litter decomposition and soil organic matter formation: a critical review. FEMS Microbiol Ecol. 2018;94:fiy175. - PubMed

