Aberrant gene activation in synovial sarcoma relies on SSX specificity and increased PRC1.1 stability
- PMID: 37735617
- PMCID: PMC10643139
- DOI: 10.1038/s41594-023-01096-3
Aberrant gene activation in synovial sarcoma relies on SSX specificity and increased PRC1.1 stability
Abstract
The SS18-SSX fusion drives oncogenic transformation in synovial sarcoma by bridging SS18, a member of the mSWI/SNF (BAF) complex, to Polycomb repressive complex 1 (PRC1) target genes. Here we show that the ability of SS18-SSX to occupy H2AK119ub1-rich regions is an intrinsic property of its SSX C terminus, which can be exploited by fusion to transcriptional regulators beyond SS18. Accordingly, SS18-SSX recruitment occurs in a manner that is independent of the core components and catalytic activity of BAF. Alternative SSX fusions are also recruited to H2AK119ub1-rich chromatin and reproduce the expression signatures of SS18-SSX by engaging with transcriptional activators. Variant Polycomb repressive complex 1.1 (PRC1.1) acts as the main depositor of H2AK119ub1 and is therefore required for SS18-SSX occupancy. Importantly, the SSX C terminus not only depends on H2AK119ub1 for localization, but also further increases it by promoting PRC1.1 complex stability. Consequently, high H2AK119ub1 levels are a feature of murine and human synovial sarcomas. These results uncover a critical role for SSX-C in mediating gene deregulation in synovial sarcoma by providing specificity to chromatin and further enabling oncofusion binding by enhancing PRC1.1 stability and H2AK119ub1 deposition.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







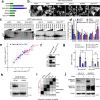


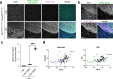

References
-
- Perry JA, Seong BKA, Stegmaier K. Biology and therapy of dominant fusion oncoproteins involving transcription factor and chromatin regulators in sarcomas. Annu. Rev. Cancer Biol. 2019;3:299–321.
-
- Clark J, et al. Identification of novel genes, SYT and SSX, involved in the t(X;18)(p11.2;q11.2) translocation found in human synovial sarcoma. Nat. Genet. 1994;7:502–508. - PubMed
-
- Ladanyi M. Fusions of the SYT and SSX genes in synovial sarcoma. Oncogene. 2001;20:5755–5762. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

