A high-field cellular DNP-supported solid-state NMR approach to study proteins with sub-cellular specificity
- PMID: 37736634
- PMCID: PMC10510770
- DOI: 10.1039/d3sc02117c
A high-field cellular DNP-supported solid-state NMR approach to study proteins with sub-cellular specificity
Abstract
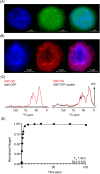
Studying the structural aspects of proteins within sub-cellular compartments is of growing interest. Dynamic nuclear polarization supported solid-state NMR (DNP-ssNMR) is uniquely suited to provide such information, but critically lacks the desired sensitivity and resolution. Here we utilize SNAPol-1, a novel biradical, to conduct DNP-ssNMR at high-magnetic fields (800 MHz/527 GHz) inside HeLa cells and isolated cell nuclei electroporated with [13C,15N] labeled ubiquitin. We report that SNAPol-1 passively diffuses and homogenously distributes within whole cells and cell nuclei providing ubiquitin spectra of high sensitivity and remarkably improved spectral resolution. For cell nuclei, physical enrichment facilitates a further 4-fold decrease in measurement time and provides an exclusive structural view of the nuclear ubiquitin pool. Taken together, these advancements enable atomic interrogation of protein conformational plasticity at atomic resolution and with sub-cellular specificity.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
The authors have no conflicts to declare.
Figures



Similar articles
-
Highly Efficient Trityl-Nitroxide Biradicals for Biomolecular High-Field Dynamic Nuclear Polarization.Chemistry. 2021 Sep 6;27(50):12758-12762. doi: 10.1002/chem.202102253. Epub 2021 Jul 22. Chemistry. 2021. PMID: 34181286
-
DNP-Supported Solid-State NMR Spectroscopy of Proteins Inside Mammalian Cells.Angew Chem Int Ed Engl. 2019 Sep 9;58(37):12969-12973. doi: 10.1002/anie.201903246. Epub 2019 Jul 19. Angew Chem Int Ed Engl. 2019. PMID: 31233270 Free PMC article.
-
A DNP-Supported Solid-State NMR Approach to Study Nucleic Acids In Situ Reveals Berberine-Stabilized Hoogsteen Structures in Mitochondria.Angew Chem Int Ed Engl. 2025 May;64(21):e202424131. doi: 10.1002/anie.202424131. Epub 2025 Mar 18. Angew Chem Int Ed Engl. 2025. PMID: 40052409 Free PMC article.
-
Is solid-state NMR enhanced by dynamic nuclear polarization?Solid State Nucl Magn Reson. 2015 Apr-May;66-67:6-20. doi: 10.1016/j.ssnmr.2015.01.003. Epub 2015 Feb 7. Solid State Nucl Magn Reson. 2015. PMID: 25779337 Review.
-
Targeted DNP for biomolecular solid-state NMR.Chem Sci. 2021 Mar 23;12(18):6223-6237. doi: 10.1039/d0sc06959k. Chem Sci. 2021. PMID: 34084422 Free PMC article. Review.
Cited by
-
Elucidating microRNA-34a organisation within human Argonaute-2 by dynamic nuclear polarisation-enhanced magic angle spinning NMR.Nucleic Acids Res. 2024 Oct 28;52(19):11995-12004. doi: 10.1093/nar/gkae744. Nucleic Acids Res. 2024. PMID: 39228364 Free PMC article.
-
In cell NMR reveals cells selectively amplify and structurally remodel amyloid fibrils.bioRxiv [Preprint]. 2024 Sep 10:2024.09.09.612142. doi: 10.1101/2024.09.09.612142. bioRxiv. 2024. PMID: 39314304 Free PMC article. Preprint.
-
Progress, Challenges and Opportunities of NMR and XL-MS for Cellular Structural Biology.JACS Au. 2024 Feb 5;4(2):369-383. doi: 10.1021/jacsau.3c00712. eCollection 2024 Feb 26. JACS Au. 2024. PMID: 38425916 Free PMC article. Review.
-
Stability of the polarization agent AsymPolPOK in intact and lysed mammalian cells.bioRxiv [Preprint]. 2024 Nov 11:2024.11.09.622814. doi: 10.1101/2024.11.09.622814. bioRxiv. 2024. Update in: J Magn Reson. 2025 May;374:107864. doi: 10.1016/j.jmr.2025.107864. PMID: 39605460 Free PMC article. Updated. Preprint.
-
Ubiquitin's Conformational Heterogeneity as Discerned by Nuclear Magnetic Resonance Spectroscopy.Chembiochem. 2024 Dec 16;25(24):e202400508. doi: 10.1002/cbic.202400508. Epub 2024 Oct 17. Chembiochem. 2024. PMID: 39140844 Free PMC article. Review.
References
-
- Cho N. H. Cheveralls K. C. Brunner A.-D. Kim K. Michaelis A. C. Raghavan P. Kobayashi H. Savy L. Li J. Y. Canaj H. Kim J. Y. S. Stewart E. M. Gnann C. McCarthy F. Cabrera J. P. Brunetti R. M. Chhun B. B. Dingle G. Hein M. Y. Huang B. Mehta S. B. Weissman J. S. Gómez-Sjöberg R. Itzhak D. N. Royer L. A. Mann M. Leonetti M. D. Science. 2022;375:eabi6983. doi: 10.1126/science.abi6983. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

