GhIMP10D, an inositol monophosphates family gene, enhances ascorbic acid and antioxidant enzyme activities to confer alkaline tolerance in Gossypium hirsutum L
- PMID: 37736713
- PMCID: PMC10515029
- DOI: 10.1186/s12870-023-04462-x
GhIMP10D, an inositol monophosphates family gene, enhances ascorbic acid and antioxidant enzyme activities to confer alkaline tolerance in Gossypium hirsutum L
Abstract
Background: Inositol monophosphates (IMP) are key enzymes in the ascorbic acid (AsA) synthesis pathways, which play vital roles in regulating plant growth and development and stresses tolerance. To date, no comprehensive analysis of the expression profile of IMP genes and their functions under abiotic stress in cotton has been reported.
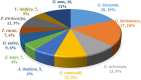
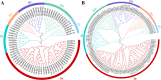
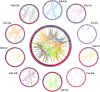
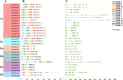
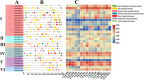
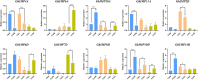
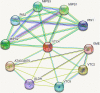
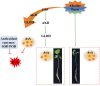
Results: In this study, the genetic characteristics, phylogenetic evolution, cis-acting elements and expression patterns of IMP gene family in cotton were systematically analyzed. A total of 28, 27, 13 and 13 IMP genes were identified in Gossypium hirsutum (G. hirsutum), Gossypium barbadense (G. barbadense), Gossypium arboreum (G. arboreum), and Gossypium raimondii (G. raimondii), respectively. Phylogenetic analysis showed that IMP family genes could cluster into 3 clades. Structure analysis of genes showed that GhIMP genes from the same subgroup had similar genetic structure and exon number. And most GhIMP family members contained hormone-related elements (abscisic acid response element, MeJA response element, gibberellin response element) and stress-related elements (low temperature response element, defense and stress response element, wound response element). After exogenous application of abscisic acid (ABA), some GhIMP genes containing ABA response elements positively responded to alkaline stress, indicating that ABA response elements played an important role in response to alkaline stress. qRT-PCR showed that most of GhIMP genes responded positively to alkaline stress, and GhIMP10D significantly upregulated under alkaline stress, with the highest up-regulated expression level. Virus-induced gene silencing (VIGS) experiment showed that compared with 156 plants, MDA content of pYL156:GhIMP10D plants increased significantly, while POD, SOD, chlorophyII and AsA content decreased significantly.
Conclusions: This study provides a thorough overview of the IMP gene family and presents a new perspective on the evolution of this gene family. In particular, some IMP genes may be involved in alkaline stress tolerance regulation, and GhIMP10D showed high expression levels in leaves, stems and roots under alkaline stress, and preliminary functional verification of GhIMP10D gene suggested that it may regulate tolerance to alkaline stress by regulating the activity of antioxidant enzymes and the content of AsA. This study contributes to the subsequent broader discussion of the structure and alkaline resistance of IMP genes in cotton.
Keywords: Alkaline tolerance; Ascorbic acid; Gossypium hirsutum L.; Inositol monophosphates.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
The authors declare no competing interests.
Figures







References
-
- Ayyub CM, Ali M, Shaheen MR, Qadri R, Khan I, Jahangir MM, Abbasi KY, Kamal S, Zain M. Enhancing the salt tolerance potential of watermelon (Citrullus lanatus) by exogenous application of salicylic acid. Am J Plant Sci. 2015;6(19):5.
-
- Munns R. Comparative physiology of salt and water stress. Plant Cell Environ. 2002;25(2):239–250. - PubMed
-
- Yang C, Chong J, Li C, Kim C, Shi D, Wang D. Osmotic adjustment and ion balance traits of an alkali resistant halophyte Kochia sieversiana during adaptation to salt and alkali conditions. Plant Soil. 2007;294(1–2):263–276.
-
- Apel K, Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol. 2004;55(1):373–399. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

