CREB1-driven CXCR4hi neutrophils promote skin inflammation in mouse models and human patients
- PMID: 37736772
- PMCID: PMC10516899
- DOI: 10.1038/s41467-023-41484-3
CREB1-driven CXCR4hi neutrophils promote skin inflammation in mouse models and human patients
Abstract
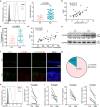
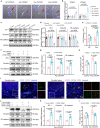
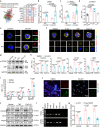
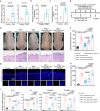
Neutrophils have a pathogenic function in inflammation via releasing pro-inflammatory mediators or neutrophil extracellular traps (NETs). However, their heterogeneity and pro-inflammatory mechanisms remain unclear. Here, we demonstrate that CXCR4hi neutrophils accumulate in the blood and inflamed skin in human psoriasis, and correlate with disease severity. Compared to CXCR4lo neutrophils, CXCR4hi neutrophils have enhanced NETs formation, phagocytic function, neutrophil degranulation, and overexpression of pro-inflammatory cytokines and chemokines in vitro. This is accompanied by a metabolic shift in CXCR4hi neutrophils toward glycolysis and lactate release, thereby promoting vascular permeability and remodeling. CXCR4 expression in neutrophils is dependent on CREB1, a transcription factor activated by TNF and CXCL12, and regulated by de novo synthesis. In vivo, CXCR4hi neutrophil infiltration amplifies skin inflammation, whereas blockade of CXCR4hi neutrophils through CXCR4 or CXCL12 inhibition leads to suppression of immune responses. In this work, our study identifies CREB1 as a critical regulator of CXCR4hi neutrophil development and characterizes the contribution of CXCR4hi neutrophils to vascular remodeling and inflammatory responses in skin.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Liew PX, Kubes P. The Neutrophil’s role during health and disease. Physiol. Rev. 2019;99:1223–1248. - PubMed
-
- Burn GL, Foti A, Marsman G, Patel DF, Zychlinsky A. The Neutrophil. Immunity. 2021;54:1377–1391. - PubMed
-
- Jarrot PA, et al. Neutrophil extracellular traps are associated with the pathogenesis of diffuse alveolar hemorrhage in murine lupus. J. Autoimmun. 2019;100:120–130. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

