Quantum biological insights into CRISPR-Cas9 sgRNA efficiency from explainable-AI driven feature engineering
- PMID: 37738140
- PMCID: PMC10602897
- DOI: 10.1093/nar/gkad736
Quantum biological insights into CRISPR-Cas9 sgRNA efficiency from explainable-AI driven feature engineering
Abstract
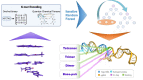
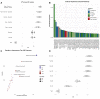
CRISPR-Cas9 tools have transformed genetic manipulation capabilities in the laboratory. Empirical rules-of-thumb have been developed for only a narrow range of model organisms, and mechanistic underpinnings for sgRNA efficiency remain poorly understood. This work establishes a novel feature set and new public resource, produced with quantum chemical tensors, for interpreting and predicting sgRNA efficiency. Feature engineering for sgRNA efficiency is performed using an explainable-artificial intelligence model: iterative Random Forest (iRF). By encoding quantitative attributes of position-specific sequences for Escherichia coli sgRNAs, we identify important traits for sgRNA design in bacterial species. Additionally, we show that expanding positional encoding to quantum descriptors of base-pair, dimer, trimer, and tetramer sequences captures intricate interactions in local and neighboring nucleotides of the target DNA. These features highlight variation in CRISPR-Cas9 sgRNA dynamics between E. coli and H. sapiens genomes. These novel encodings of sgRNAs enhance our understanding of the elaborate quantum biological processes involved in CRISPR-Cas9 machinery.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


Similar articles
-
sgRNA Sequence Motifs Blocking Efficient CRISPR/Cas9-Mediated Gene Editing.Cell Rep. 2019 Jan 29;26(5):1098-1103.e3. doi: 10.1016/j.celrep.2019.01.024. Cell Rep. 2019. PMID: 30699341 Free PMC article.
-
Unpredicted central inversion in a sgRNA flanked by inverted repeats.Mol Biol Rep. 2020 Aug;47(8):6375-6378. doi: 10.1007/s11033-020-05524-1. Epub 2020 May 18. Mol Biol Rep. 2020. PMID: 32424520
-
Selection of highly efficient sgRNAs for CRISPR/Cas9-based plant genome editing.Sci Rep. 2016 Feb 19;6:21451. doi: 10.1038/srep21451. Sci Rep. 2016. PMID: 26891616 Free PMC article.
-
Review of CRISPR/Cas9 sgRNA Design Tools.Interdiscip Sci. 2018 Jun;10(2):455-465. doi: 10.1007/s12539-018-0298-z. Epub 2018 Apr 11. Interdiscip Sci. 2018. PMID: 29644494 Review.
-
Optimization Strategies for the CRISPR-Cas9 Genome-Editing System.Cold Spring Harb Protoc. 2016 Oct 3;2016(10). doi: 10.1101/pdb.top090894. Cold Spring Harb Protoc. 2016. PMID: 27698246 Review.
Cited by
-
Advancing CRISPR base editing technology through innovative strategies and ideas.Sci China Life Sci. 2025 Mar;68(3):610-627. doi: 10.1007/s11427-024-2699-5. Epub 2024 Sep 2. Sci China Life Sci. 2025. PMID: 39231901 Review.
-
Application of functional genomics for domestication of novel non-model microbes.J Ind Microbiol Biotechnol. 2024 Jan 9;51:kuae022. doi: 10.1093/jimb/kuae022. J Ind Microbiol Biotechnol. 2024. PMID: 38925657 Free PMC article. Review.
-
Transitioning from wet lab to artificial intelligence: a systematic review of AI predictors in CRISPR.J Transl Med. 2025 Feb 4;23(1):153. doi: 10.1186/s12967-024-06013-w. J Transl Med. 2025. PMID: 39905452 Free PMC article.
-
Research Hotspots and Trends of NK Cell Immunotherapy for Acute Myeloid Leukemia: A Bibliometric Analysis From 2000 to 2023.Cancer Control. 2024 Jan-Dec;31:10732748241310937. doi: 10.1177/10732748241310937. Cancer Control. 2024. PMID: 39703189 Free PMC article. Review.
References
-
- Doudna J.A., Charpentier E.. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014; 346:1258096. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

