Measuring prion propagation in single bacteria elucidates a mechanism of loss
- PMID: 37738299
- PMCID: PMC10523482
- DOI: 10.1073/pnas.2221539120
Measuring prion propagation in single bacteria elucidates a mechanism of loss
Abstract
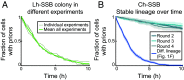
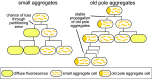
Prions are self-propagating protein aggregates formed by specific proteins that can adopt alternative folds. Prions were discovered as the cause of the fatal transmissible spongiform encephalopathies in mammals, but prions can also constitute nontoxic protein-based elements of inheritance in fungi and other species. Prion propagation has recently been shown to occur in bacteria for more than a hundred cell divisions, yet a fraction of cells in these lineages lost the prion through an unknown mechanism. Here, we investigate prion propagation in single bacterial cells as they divide using microfluidics and fluorescence microscopy. We show that the propagation occurs in two distinct modes. In a fraction of the population, cells had multiple small visible aggregates and lost the prion through random partitioning of aggregates to one of the two daughter cells at division. In the other subpopulation, cells had a stable large aggregate localized to the pole; upon division the mother cell retained this polar aggregate and a daughter cell was generated that contained small aggregates. Extending our findings to prion domains from two orthologous proteins, we observe similar propagation and loss properties. Our findings also provide support for the suggestion that bacterial prions can form more than one self-propagating state. We implement a stochastic version of the molecular model of prion propagation from yeast and mammals that recapitulates all the observed single-cell properties. This model highlights challenges for prion propagation that are unique to prokaryotes and illustrates the conservation of fundamental characteristics of prion propagation.
Keywords: Escherichia coli; microfluidics; prions; protein-based heredity; single-cell microscopy.
Conflict of interest statement
The authors declare no competing interest.
Figures




Update of
-
Measuring prion propagation in single bacteria elucidates mechanism of loss.bioRxiv [Preprint]. 2023 Jan 12:2023.01.11.523042. doi: 10.1101/2023.01.11.523042. bioRxiv. 2023. Update in: Proc Natl Acad Sci U S A. 2023 Sep 26;120(39):e2221539120. doi: 10.1073/pnas.2221539120. PMID: 36712035 Free PMC article. Updated. Preprint.
References
-
- Prusiner S. B., Novel proteinaceous infectious particles cause scrapie. Science 216, 136–144 (1982). - PubMed
-
- Glover J. R., et al. , Self-Seeded fibers formed by sup35, the protein determinant of [PSI+], a heritable prion-like factor of s. cerevisiae. Cell 89, 811–819 (1997). - PubMed
-
- True H. L., Lindquist S. L., A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature 407, 477–483 (2000). - PubMed
-
- Levkovich S. A., Rencus-Lazar S., Gazit E., Bar-Yosef D. L., Microbial prions: Dawn of a new era. Trends Biochem. Sci. 46, 391–405 (2021). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

