Genome-wide variation in the Angolan Namib Desert reveals unique pre-Bantu ancestry
- PMID: 37738339
- PMCID: PMC10516492
- DOI: 10.1126/sciadv.adh3822
Genome-wide variation in the Angolan Namib Desert reveals unique pre-Bantu ancestry
Abstract
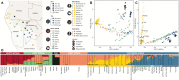
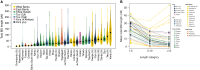
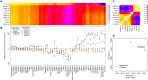
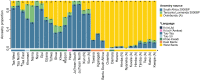
Ancient DNA studies reveal the genetic structure of Africa before the expansion of Bantu-speaking agriculturalists; however, the impact of now extinct hunter-gatherer and herder societies on the genetic makeup of present-day African groups remains elusive. Here, we uncover the genetic legacy of pre-Bantu populations from the Angolan Namib Desert, where we located small-scale groups associated with enigmatic forager traditions, as well as the last speakers of the Khoe-Kwadi family's Kwadi branch. By applying an ancestry decomposition approach to genome-wide data from these and other African populations, we reconstructed the fine-scale histories of contact emerging from the migration of Khoe-Kwadi-speaking pastoralists and identified a deeply divergent ancestry, which is exclusively shared between groups from the Angolan Namib and adjacent areas of Namibia. The unique genetic heritage of the Namib peoples shows how modern DNA research targeting understudied regions of high ethnolinguistic diversity can complement ancient DNA studies in probing the deep genetic structure of the African continent.
Figures
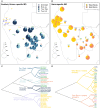
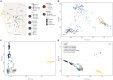
References
-
- J. K. Pickrell, N. Patterson, C. Barbieri, F. Berthold, L. Gerlach, T. Güldemann, B. Kure, S. W. Mpoloka, H. Nakagawa, C. Naumann, M. Lipson, P. R. Loh, J. Lachance, J. Mountain, C. D. Bustamante, B. Berger, S. A. Tishkoff, B. M. Henn, M. Stoneking, D. Reich, B. Pakendorf, The genetic prehistory of southern Africa. Nat. Commun. 3, 1143 (2012). - PMC - PubMed
-
- T. Güldemann, A linguist’s view: Khoe-Kwadi speakers as the earliest food-producers of southern Africa. Southern African Humanit. 20, 99–132 (2008).
-
- E. O. J. Westphal, The age of “Bushman” languages in southern African pre-history, in Bushman and Hottentot Linguistic Studies, J. W. Snyman, Ed. (University of South Africa, 1980), pp. 59–79.

