Metabolic engineering and optimization of Escherichia coli co-culture for the de novo synthesis of genkwanin
- PMID: 37738435
- PMCID: PMC10565888
- DOI: 10.1093/jimb/kuad030
Metabolic engineering and optimization of Escherichia coli co-culture for the de novo synthesis of genkwanin
Abstract
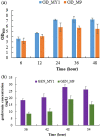
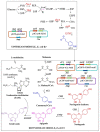
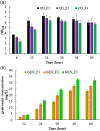
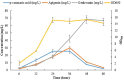
Genkwanin has various significant roles in nutrition, biomedicine, and pharmaceutical biology. Previously, this compound was chiefly produced by plant-originated extraction or chemical synthesis. However, due to increasing concern and demand for safe food and environmental issues, the biotechnological production of genkwanin and other bioactive compounds based on safe, cheap, and renewable substrates has gained much interest. This paper described recombinant Escherichia coli-based co-culture engineering that was reconstructed for the de novo production of genkwanin from d-glucose. The artificial genkwanin biosynthetic chain was divided into 2 modules in which the upstream strain contained the genes for synthesizing p-coumaric acid from d-glucose, and the downstream module contained a gene cluster that produced the precursor apigenin and the final product, genkwanin. The Box-Behnken design, a response surface methodology, was used to empirically model the production of genkwanin and optimize its productivity. As a result, the application of the designed co-culture improved the genkwanin production by 48.8 ± 1.3 mg/L or 1.7-fold compared to the monoculture. In addition, the scale-up of genkwanin bioproduction by a bioreactor resulted in 68.5 ± 1.9 mg/L at a 48 hr time point. The combination of metabolic engineering and fermentation technology was therefore a very efficient and applicable approach to enhance the production of other bioactive compounds.
Keywords: De novo production; Escherichia coli; Co-culture; Genkwanin; Metabolic engineering.
© The Author(s) 2023. Published by Oxford University Press on behalf of Society of Industrial Microbiology and Biotechnology.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures



References
-
- Adams A. M., Kaplan N. A., Wei Z., Brinton J. D., Monnier C. S., Enacopol A. L., Ramelot T. A., Jones J. A. (2019). In vivo production of psilocybin in E. coli. Metabolic Engineering, 56, 111–119. - PubMed
-
- Cui H., Song M. C., Ban Y. H., Jun S. Y., Kwon A. S., Lee J. Y., Yoon Y. J. (2019). High-yield production of multiple O-methylated phenylpropanoids by the engineered Escherichia coli–Streptomyces cocultivation system. Microbial Cell Factories, 18(1), 67. 10.1186/s12934-019-1118-9 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

