Mass Spectrometry-Based Proteogenomics: New Therapeutic Opportunities for Precision Medicine
- PMID: 37738504
- PMCID: PMC10950354
- DOI: 10.1146/annurev-pharmtox-022723-113921
Mass Spectrometry-Based Proteogenomics: New Therapeutic Opportunities for Precision Medicine
Abstract
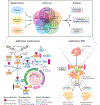
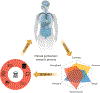
Proteogenomics refers to the integration of comprehensive genomic, transcriptomic, and proteomic measurements from the same samples with the goal of fully understanding the regulatory processes converting genotypes to phenotypes, often with an emphasis on gaining a deeper understanding of disease processes. Although specific genetic mutations have long been known to drive the development of multiple cancers, gene mutations alone do not always predict prognosis or response to targeted therapy. The benefit of proteogenomics research is that information obtained from proteins and their corresponding pathways provides insight into therapeutic targets that can complement genomic information by providing an additional dimension regarding the underlying mechanisms and pathophysiology of tumors. This review describes the novel insights into tumor biology and drug resistance derived from proteogenomic analysis while highlighting the clinical potential of proteogenomic observations and advances in technique and analysis tools.
Keywords: drug resistance; oncoproteomics; phosphoproteomics; proteogenomics; single-cell proteomics.
Figures


References
-
- Druker BJ, Tamura S, Buchdunger E, Ohno S, Segal GM, et al. 1996. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat. Med 2:561–66 - PubMed
-
- Druker BJ, Talpaz M, Resta DJ, Peng B, Buchdunger E, et al. 2001. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N. Engl. J. Med 344:1031–37 - PubMed
-
- Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, et al. 2001. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N. Engl. J. Med 344:783–92 - PubMed
-
- Wang Z, Jensen MA, Zenklusen JC. 2016. A practical guide to The Cancer Genome Atlas (TCGA). Methods Mol. Biol 1418:111–41 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

