Identification and application of a candidate gene AhAftr1 for aflatoxin production resistance in peanut seed (Arachis hypogaea L.)
- PMID: 37739123
- PMCID: PMC11331177
- DOI: 10.1016/j.jare.2023.09.014
Identification and application of a candidate gene AhAftr1 for aflatoxin production resistance in peanut seed (Arachis hypogaea L.)
Abstract
Introduction: Peanut is susceptible to infection of Aspergillus fungi and conducive to aflatoxin contamination, hence developing aflatoxin-resistant variety is highly meaningful. Identifying functional genes or loci conferring aflatoxin resistance and molecular diagnostic marker are crucial for peanut breeding.
Objectives: This work aims to (1) identify candidate gene for aflatoxin production resistance, (2) reveal the related resistance mechanism, and (3) develop diagnostic marker for resistance breeding program.
Methods: Resistance to aflatoxin production in a recombined inbred line (RIL) population derived from a high-yielding variety Xuhua13 crossed with an aflatoxin-resistant genotype Zhonghua 6 was evaluated under artificial inoculation for three consecutive years. Both genetic linkage analysis and QTL-seq were conducted for QTL mapping. The candidate gene was further fine-mapped using a secondary segregation mapping population and validated by transgenic experiments. RNA-Seq analysis among resistant and susceptible RILs was used to reveal the resistance pathway for the candidate genes.
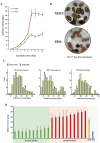
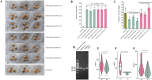
Results: The major effect QTL qAFTRA07.1 for aflatoxin production resistance was mapped to a 1.98 Mbp interval. A gene, AhAftr1 (Arachis hypogaea Aflatoxin resistance 1), was detected structure variation (SV) in leucine rich repeat (LRR) domain of its production, and involved in disease resistance response through the effector-triggered immunity (ETI) pathway. Transgenic plants with overexpression of AhAftr1(ZH6) exhibited 57.3% aflatoxin reduction compared to that of AhAftr1(XH13). A molecular diagnostic marker AFTR.Del.A07 was developed based on the SV. Thirty-six lines, with aflatoxin content decrease by over 77.67% compared to the susceptible control Zhonghua12 (ZH12), were identified from a panel of peanut germplasm accessions and breeding lines through using AFTR.Del.A07.
Conclusion: Our findings would provide insights of aflatoxin production resistance mechanisms and laid meaningful foundation for further breeding programs.
Keywords: Aflatoxin production resistance; Diagnositic marker; ETI; NB-LRRs; Peanut.
Copyright © 2024. Production and hosting by Elsevier B.V.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures










Similar articles
-
Emerging Strategies for Aflatoxin Resistance in Peanuts via Precision Breeding.Toxins (Basel). 2025 Aug 6;17(8):394. doi: 10.3390/toxins17080394. Toxins (Basel). 2025. PMID: 40864070 Free PMC article. Review.
-
Identification of candidate genes associated with resistance to aflatoxin production in peanut through genetic mapping and transcriptome analysis.Theor Appl Genet. 2025 Mar 13;138(4):71. doi: 10.1007/s00122-025-04822-1. Theor Appl Genet. 2025. PMID: 40074866
-
Identification of genomic regions and diagnostic markers for resistance to aflatoxin contamination in peanut (Arachis hypogaea L.).BMC Genet. 2019 Mar 12;20(1):32. doi: 10.1186/s12863-019-0734-z. BMC Genet. 2019. PMID: 30866805 Free PMC article.
-
High-density SNP map facilitates fine mapping of QTLs and candidate genes discovery for Aspergillus flavus resistance in peanut (Arachis hypogaea).Theor Appl Genet. 2020 Jul;133(7):2239-2257. doi: 10.1007/s00122-020-03594-0. Epub 2020 Apr 13. Theor Appl Genet. 2020. PMID: 32285164
-
BSA‑seq and genetic mapping reveals AhRt2 as a candidate gene responsible for red testa of peanut.Theor Appl Genet. 2022 May;135(5):1529-1540. doi: 10.1007/s00122-022-04051-w. Epub 2022 Feb 15. Theor Appl Genet. 2022. PMID: 35166897 Review.
Cited by
-
Emerging Strategies for Aflatoxin Resistance in Peanuts via Precision Breeding.Toxins (Basel). 2025 Aug 6;17(8):394. doi: 10.3390/toxins17080394. Toxins (Basel). 2025. PMID: 40864070 Free PMC article. Review.
-
Identification of candidate genes associated with resistance to aflatoxin production in peanut through genetic mapping and transcriptome analysis.Theor Appl Genet. 2025 Mar 13;138(4):71. doi: 10.1007/s00122-025-04822-1. Theor Appl Genet. 2025. PMID: 40074866
-
Integration of BSA-seq and high-resolution mapping reveals genomic regions and candidate genes controlling seed oil accumulation in peanut (Arachis hypogaea L.).Theor Appl Genet. 2025 Jun 17;138(7):154. doi: 10.1007/s00122-025-04939-3. Theor Appl Genet. 2025. PMID: 40528052
References
-
- Payne G.A., Widstrom N.W. Aflatoxin in maize. Crit Rev Plant Sci. 1992;10(5):423–440. doi: 10.1080/07352689209382320. - DOI
-
- Kurtzman C.P., Smiley M.J., Robnett C.J., Wicklow D.T. DNA relatedness among wild and domesticated species in the Aspergillus flavus group. Mycologia. 1986;78(6):955–959. doi: 10.1080/00275514.1986.12025355. - DOI
-
- Bhatnagar-Mathur P., Sunkara S., Bhatnagar-Panwar M., Waliyar F., Sharma K.K. Biotechnological advances for combating Aspergillus flavus and aflatoxin contamination in crops. Plant science : an international journal of experimental plant biology. 2015;234:119–132. doi: 10.1016/j.plantsci.2015.02.009. - DOI - PubMed
-
- Holbrook C.C., Kvien C.K., Rucker K.S., Wilson D.M., Matheron M.E. Preharvest aflatoxin contamination in drought-tolerant and drought-intolerant peanut genotypes. Peanut Science. 2000;27(2):45–48.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

