Characterization and genomic analysis of Stutzerimonas stutzeri phage vB_PstS_ZQG1, representing a novel viral genus
- PMID: 37739268
- PMCID: PMC10520572
- DOI: 10.1016/j.virusres.2023.199226
Characterization and genomic analysis of Stutzerimonas stutzeri phage vB_PstS_ZQG1, representing a novel viral genus
Abstract
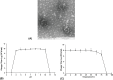
Stutzerimonas stutzeri is an opportunistic pathogenic bacterium belonging to the Gammaproteobacteria, exhibiting wide distribution in the environment and playing significant ecological roles such as nitrogen fixation or pollutant degradation. Despite its ecological importance, only two S. stutzeri phages have been isolated to date. Here, a novel S. stutzeri phage, vB_PstS_ZQG1, was isolated from the surface seawater of Qingdao, China. Transmission electron microscopy analysis indicates that vB_PstS_ZQG1 has a morphology characterized by a long non-contractile tail. The genomic sequence of vB_PstS_ZQG1 contains a linear, double-strand 61,790-bp with the G+C content of 53.24% and encodes 90 putative open reading frames. Two auxiliary metabolic genes encoding TolA protein and nucleotide pyrophosphohydrolase were identified, which are likely involved in host adaptation and phage reproduction. Phylogenetic and comparative genomic analyses demonstrated that vB_PstS_ZQG1 exhibits low similarity with previously isolated phages or uncultured viruses (average nucleotide identity values range from 21.7 to 29.4), suggesting that it represents a novel viral genus by itself, here named as Fuevirus. Biogeographic analysis showed that vB_PstS_ZQG1 was only detected in epipelagic and mesopelagic zone with low abundance. In summary, our findings of the phage vB_PstS_ZQG1 will provide helpful insights for further research on the interactions between S. stutzeri phages and their hosts, and contribute to discovering unknown viral sequences in the metagenomic database.
Keywords: Biogeographic analysis; Fuevirus; Genomic and phylogenetic analysis; Stutzerimonas stutzeri; Vb_psts_zqg1.
Copyright © 2023 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures





Similar articles
-
Characterization and genomic analysis of a novel lytic phage vB_PstM_ZRG1 infecting Stutzerimonas stutzeri, representing a new viral genus, Elithevirus.Virus Res. 2023 Sep;334:199183. doi: 10.1016/j.virusres.2023.199183. Epub 2023 Jul 28. Virus Res. 2023. PMID: 37499764 Free PMC article.
-
Psychrobacter Phage Encoding an Antibiotics Resistance Gene Represents a Novel Caudoviral Family.Microbiol Spectr. 2023 Aug 17;11(4):e0533522. doi: 10.1128/spectrum.05335-22. Epub 2023 Jun 5. Microbiol Spectr. 2023. PMID: 37272818 Free PMC article.
-
Shewanella phage encoding a putative anti-CRISPR-like gene represents a novel potential viral family.Microbiol Spectr. 2024 Feb 6;12(2):e0336723. doi: 10.1128/spectrum.03367-23. Epub 2024 Jan 12. Microbiol Spectr. 2024. PMID: 38214523 Free PMC article.
-
Genome and Ecology of a Novel Alteromonas Podovirus, ZP6, Representing a New Viral Genus, Mareflavirus.Microbiol Spectr. 2021 Oct 31;9(2):e0046321. doi: 10.1128/Spectrum.00463-21. Epub 2021 Oct 13. Microbiol Spectr. 2021. PMID: 34643440 Free PMC article.
-
A novel flavobacterial phage abundant during green tide, representing a new viral family, Zblingviridae.Appl Environ Microbiol. 2024 Jul 24;90(7):e0036724. doi: 10.1128/aem.00367-24. Epub 2024 Jul 2. Appl Environ Microbiol. 2024. PMID: 38953371 Free PMC article.
Cited by
-
Characterization and genome-informatic analysis of a novel lytic mendocina phage vB_PmeS_STP12 suitable for phage therapy pseudomonas or biocontrol.Mol Biol Rep. 2024 Mar 14;51(1):419. doi: 10.1007/s11033-024-09362-3. Mol Biol Rep. 2024. PMID: 38483683
-
Isolation and characterization of a novel lytic bacteriophage Pv27 with biocontrol potential against Vibrio parahaemolyticus infections in shrimp.PeerJ. 2025 May 6;13:e19421. doi: 10.7717/peerj.19421. eCollection 2025. PeerJ. 2025. PMID: 40352283 Free PMC article.
-
Pangenomic landscapes shape performances of a synthetic genetic circuit across Stutzerimonas species.mSystems. 2024 Sep 17;9(9):e0084924. doi: 10.1128/msystems.00849-24. Epub 2024 Aug 21. mSystems. 2024. PMID: 39166875 Free PMC article.
References
-
- Angly F., Youle M., Nosrat B., Srinagesh S., Rodriguez-Brito B., McNairnie P., Deyanat-Yazdi G., Breitbart M., Rohwer F. Genomic analysis of multiple Roseophage SIO1 strains. Environ. Microbiol. 2009;11:2863–2873. - PubMed
-
- Bastian M., Heymann S., Jacomy M. Proceedings of the international AAAI conference on web and social media. 2009. Gephi: an open source software for exploring and manipulating networks; pp. 361–362.
-
- Bin Jang H., Bolduc B., Zablocki O., Kuhn J.H., Roux S., Adriaenssens E.M., Brister J.R., Kropinski A.M., Krupovic M., Lavigne R., Turner D., Sullivan M.B. Taxonomic assignment of uncultivated prokaryotic virus genomes is enabled by gene-sharing networks. Nat. Biotechnol. 2019;37:632–639. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

