High-throughput, fluorescent-aptamer-based measurements of steady-state transcription rates for the Mycobacterium tuberculosis RNA polymerase
- PMID: 37739412
- PMCID: PMC10602862
- DOI: 10.1093/nar/gkad761
High-throughput, fluorescent-aptamer-based measurements of steady-state transcription rates for the Mycobacterium tuberculosis RNA polymerase
Abstract
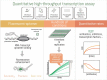
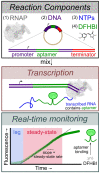
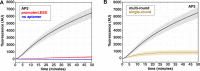
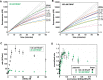
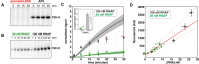
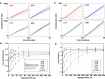
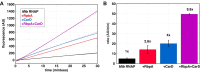
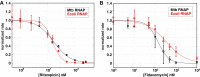
The first step in gene expression is the transcription of DNA sequences into RNA. Regulation at the level of transcription leads to changes in steady-state concentrations of RNA transcripts, affecting the flux of downstream functions and ultimately cellular phenotypes. Changes in transcript levels are routinely followed in cellular contexts via genome-wide sequencing techniques. However, in vitro mechanistic studies of transcription have lagged with respect to throughput. Here, we describe the use of a real-time, fluorescent-aptamer-based method to quantitate steady-state transcription rates of the Mycobacterium tuberculosis RNA polymerase. We present clear controls to show that the assay specifically reports on promoter-dependent, full-length RNA transcription rates that are in good agreement with the kinetics determined by gel-resolved, α-32P NTP incorporation experiments. We illustrate how the time-dependent changes in fluorescence can be used to measure regulatory effects of nucleotide concentrations and identity, RNAP and DNA concentrations, transcription factors, and antibiotics. Our data showcase the ability to easily perform hundreds of parallel steady-state measurements across varying conditions with high precision and reproducibility to facilitate the study of the molecular mechanisms of bacterial transcription.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

Update of
-
High-throughput, fluorescent-aptamer-based measurements of steady-state transcription rates for Mycobacterium tuberculosis RNA polymerase.bioRxiv [Preprint]. 2023 Mar 13:2023.03.13.532464. doi: 10.1101/2023.03.13.532464. bioRxiv. 2023. Update in: Nucleic Acids Res. 2023 Oct 27;51(19):e99. doi: 10.1093/nar/gkad761. PMID: 36993414 Free PMC article. Updated. Preprint.
Similar articles
-
High-throughput, fluorescent-aptamer-based measurements of steady-state transcription rates for Mycobacterium tuberculosis RNA polymerase.bioRxiv [Preprint]. 2023 Mar 13:2023.03.13.532464. doi: 10.1101/2023.03.13.532464. bioRxiv. 2023. Update in: Nucleic Acids Res. 2023 Oct 27;51(19):e99. doi: 10.1093/nar/gkad761. PMID: 36993414 Free PMC article. Updated. Preprint.
-
CarD and RbpA modify the kinetics of initial transcription and slow promoter escape of the Mycobacterium tuberculosis RNA polymerase.Nucleic Acids Res. 2019 Jul 26;47(13):6685-6698. doi: 10.1093/nar/gkz449. Nucleic Acids Res. 2019. PMID: 31127308 Free PMC article.
-
The calculation of transcript flux ratios reveals single regulatory mechanisms capable of activation and repression.Proc Natl Acad Sci U S A. 2018 Dec 11;115(50):E11604-E11613. doi: 10.1073/pnas.1809454115. Epub 2018 Nov 21. Proc Natl Acad Sci U S A. 2018. PMID: 30463953 Free PMC article.
-
RNA capping by mitochondrial and multi-subunit RNA polymerases.Transcription. 2018;9(5):292-297. doi: 10.1080/21541264.2018.1456258. Epub 2018 Apr 25. Transcription. 2018. PMID: 29624107 Free PMC article. Review.
-
Transcriptional Pausing as a Mediator of Bacterial Gene Regulation.Annu Rev Microbiol. 2021 Oct 8;75:291-314. doi: 10.1146/annurev-micro-051721-043826. Epub 2021 Aug 4. Annu Rev Microbiol. 2021. PMID: 34348029 Review.
Cited by
-
Single-stranded DNA binding to the transcription factor PafBC triggers the mycobacterial DNA damage response.Sci Adv. 2025 Feb 7;11(6):eadq9054. doi: 10.1126/sciadv.adq9054. Epub 2025 Feb 7. Sci Adv. 2025. PMID: 39919186 Free PMC article.
-
Quantitative real-time in vitro transcription assay (QRIVTA) for transcriptional regulation studies.Protein Cell. 2025 Jun 20;16(6):484-490. doi: 10.1093/procel/pwae054. Protein Cell. 2025. PMID: 39387643 Free PMC article. No abstract available.
-
Regulation of steady state ribosomal transcription in Mycobacterium tuberculosis: Intersection of sigma subunits, superhelicity, and transcription factors.J Biol Chem. 2025 Jun 12;301(8):110369. doi: 10.1016/j.jbc.2025.110369. Online ahead of print. J Biol Chem. 2025. PMID: 40516872 Free PMC article.
-
Regulation of Steady State Ribosomal Transcription in Mycobacterium tuberculosis: Intersection of Sigma Subunits, Superhelicity, and Transcription Factors.bioRxiv [Preprint]. 2025 Feb 27:2025.02.24.639987. doi: 10.1101/2025.02.24.639987. bioRxiv. 2025. Update in: J Biol Chem. 2025 Jun 12;301(8):110369. doi: 10.1016/j.jbc.2025.110369. PMID: 40060575 Free PMC article. Updated. Preprint.
References
-
- McClure W.R. Mechanism and control of transcription initiation in prokaryotes. Annu. Rev. Biochem. 1985; 54:171–204. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

