SCARB2 drives hepatocellular carcinoma tumor initiating cells via enhanced MYC transcriptional activity
- PMID: 37739936
- PMCID: PMC10517016
- DOI: 10.1038/s41467-023-41593-z
SCARB2 drives hepatocellular carcinoma tumor initiating cells via enhanced MYC transcriptional activity
Abstract
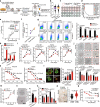
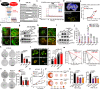
CSCs (Cancer stem cells) with distinct metabolic features are considered to cause HCC (hepatocellular carcinoma) initiation, metastasis and therapeutic resistance. Here, we perform a metabolic gene CRISPR/Cas9 knockout library screen in tumorspheres derived from HCC cells and find that deletion of SCARB2 suppresses the cancer stem cell-like properties of HCC cells. Knockout of Scarb2 in hepatocytes attenuates HCC initiation and progression in both MYC-driven and DEN (diethylnitrosamine)-induced HCC mouse models. Mechanistically, binding of SCARB2 with MYC promotes MYC acetylation by interfering with HDCA3-mediated MYC deacetylation on lysine 148 and subsequently enhances MYC transcriptional activity. Screening of a database of FDA (Food and Drug Administration)-approved drugs shows Polymyxin B displays high binding affinity for SCARB2 protein, disrupts the SCARB2-MYC interaction, decreases MYC activity, and reduces the tumor burden. Our study identifies SCARB2 as a functional driver of HCC and suggests Polymyxin B-based treatment as a targeted therapeutic option for HCC.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

