Guadecitabine plus ipilimumab in unresectable melanoma: five-year follow-up and integrated multi-omic analysis in the phase 1b NIBIT-M4 trial
- PMID: 37739939
- PMCID: PMC10516894
- DOI: 10.1038/s41467-023-40994-4
Guadecitabine plus ipilimumab in unresectable melanoma: five-year follow-up and integrated multi-omic analysis in the phase 1b NIBIT-M4 trial
Abstract
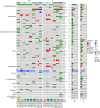
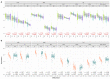
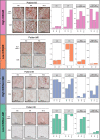
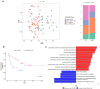
Association with hypomethylating agents is a promising strategy to improve the efficacy of immune checkpoint inhibitors-based therapy. The NIBIT-M4 was a phase Ib, dose-escalation trial in patients with advanced melanoma of the hypomethylating agent guadecitabine combined with the anti-CTLA-4 antibody ipilimumab that followed a traditional 3 + 3 design (NCT02608437). Patients received guadecitabine 30, 45 or 60 mg/m2/day subcutaneously on days 1 to 5 every 3 weeks starting on week 0 for a total of four cycles, and ipilimumab 3 mg/kg intravenously starting on day 1 of week 1 every 3 weeks for a total of four cycles. Primary outcomes of safety, tolerability, and maximum tolerated dose of treatment were previously reported. Here we report the 5-year clinical outcome for the secondary endpoints of overall survival, progression free survival, and duration of response, and an exploratory integrated multi-omics analysis on pre- and on-treatment tumor biopsies. With a minimum follow-up of 45 months, the 5-year overall survival rate was 28.9% and the median duration of response was 20.6 months. Re-expression of immuno-modulatory endogenous retroviruses and of other repetitive elements, and a mechanistic signature of guadecitabine are associated with response. Integration of a genetic immunoediting index with an adaptive immunity signature stratifies patients/lesions into four distinct subsets and discriminates 5-year overall survival and progression free survival. These results suggest that coupling genetic immunoediting with activation of adaptive immunity is a relevant requisite for achieving long term clinical benefit by epigenetic immunomodulation in advanced melanoma patients.
© 2023. Springer Nature Limited.
Conflict of interest statement
A.M.D.G. has served as consultant and/or advisor to Incyte, Pierre Fabre, Glaxo Smith Kline, Bristol-Myers Squibb, Merck Sharp Dohme, and Sanofi and has received compensated educational activities from Bristol-Myers Squibb, Merck Sharp Dohme, Pierre Fabre, and Sanofi. W.H.F. has served as consultant and/or advisor to AstraZeneca, Adaptimmune, Catalym, OOSE Immunotherapeutics, and Novartis, and reports receiving speakers bureau honoraria from Bristol-Myers Squibb. MM has served as consultant and/or advisor to Roche, Bristol-Myers Squibb, Merck Sharp Dohme, Incyte, AstraZeneca, Amgen, Pierre Fabre, Eli Lilly, Glaxo Smith Kline, Sciclone, Sanofi, Alfasigma, and Merck Serono; and own shares in Theravance and Epigen Therapeutics, Srl. MC serves as consultant and/or advisor to Moderna Therapeutics and is the founder and owns shares of Immunomica srl. Other authors have nothing to declare.
Figures




References
-
- Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348:56–61. - PubMed
-
- Paz-Ares L, et al. Durvalumab plus platinum-etoposide versus platinum-etoposide in first-line treatment of extensive-stage small-cell lung cancer (CASPIAN): a randomised, controlled, open-label, phase 3 trial. Lancet. 2019;394:1929–1939. - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical

