Camel nanobody-based B7-H3 CAR-T cells show high efficacy against large solid tumours
- PMID: 37739951
- PMCID: PMC10517151
- DOI: 10.1038/s41467-023-41631-w
Camel nanobody-based B7-H3 CAR-T cells show high efficacy against large solid tumours
Abstract
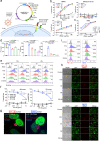
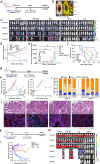
Rational design of chimeric antigen receptor T (CAR-T) cells based on the recognition of antigenic epitopes capable of evoking the most potent CAR activation is an important objective in optimizing immune therapy. In solid tumors, the B7-H3 transmembrane protein is an emerging target that harbours two distinct epitope motifs, IgC and IgV, in its ectodomain. Here, we generate dromedary camel nanobodies targeting B7-H3 and demonstrate that CAR-T cells, based on the nanobodies recognizing the IgC but not IgV domain, had potent antitumour activity against large tumors in female mice. These CAR-T cells are characterized by highly activated T cell signaling and significant tumor infiltration. Single-cell transcriptome RNA sequencing coupled with functional T-cell proteomics analysis uncovers the top-upregulated genes that might be critical for the persistence of polyfunctional CAR-T cells in mice. Our results highlight the importance of the specific target antigen epitope in governing optimal CAR-T activity and provide a nanobody-based B7-H3 CAR-T product for use in solid tumor therapy.
© 2023. Springer Nature Limited.
Conflict of interest statement
M.H., R.W., B.S.C., and D.L. are inventors on international patent application no. PCT/US2020/056601 (WO/2021/081052) assigned to the NIH, “High affinity nanobodies targeting B7-H3 (CD276) for treating multiple solid tumors”. W.N., J.Z., and S.M. are employed by and have equity ownership in IsoPlexis. All authors declare no other competing interests.
Figures





References
-
- Castellarin M, Watanabe K, June CH, Kloss CC, Posey AD. Driving cars to the clinic for solid tumors. Gene Ther. 2018;25:165–175. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

