Acetylation discriminates disease-specific tau deposition
- PMID: 37739953
- PMCID: PMC10517010
- DOI: 10.1038/s41467-023-41672-1
Acetylation discriminates disease-specific tau deposition
Abstract
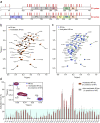
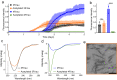
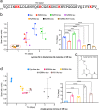
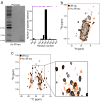
Pathogenic aggregation of the protein tau is a hallmark of Alzheimer's disease and several other tauopathies. Tauopathies are characterized by the deposition of specific tau isoforms as disease-related tau filament structures. The molecular processes that determine isoform-specific deposition of tau are however enigmatic. Here we show that acetylation of tau discriminates its isoform-specific aggregation. We reveal that acetylation strongly attenuates aggregation of four-repeat tau protein, but promotes amyloid formation of three-repeat tau. We further identify acetylation of lysine 298 as a hot spot for isoform-specific tau aggregation. Solid-state NMR spectroscopy demonstrates that amyloid fibrils formed by unmodified and acetylated three-repeat tau differ in structure indicating that site-specific acetylation modulates tau structure. The results implicate acetylation as a critical regulator that guides the selective aggregation of three-repeat tau and the development of tau isoform-specific neurodegenerative diseases.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Wang Y, Mandelkow E. Tau in physiology and pathology. Nat. Rev. Neurosci. 2016;17:5–21. - PubMed
-
- Li D, Liu C. Hierarchical chemical determination of amyloid polymorphs in neurodegenerative disease. Nat. Chem. Biol. 2021;17:237–245. - PubMed
-
- Andreadis A. Misregulation of tau alternative splicing in neurodegeneration and dementia. Prog. Mol. Subcell. Biol. 2006;44:89–107. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

