Viral maintenance and excretion dynamics of coronaviruses within an Egyptian rousette fruit bat maternal colony: considerations for spillover
- PMID: 37739999
- PMCID: PMC10517123
- DOI: 10.1038/s41598-023-42938-w
Viral maintenance and excretion dynamics of coronaviruses within an Egyptian rousette fruit bat maternal colony: considerations for spillover
Abstract
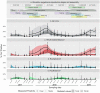
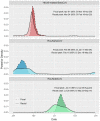
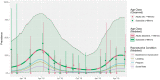
Novel coronavirus species of public health and veterinary importance have emerged in the first two decades of the twenty-first century, with bats identified as natural hosts for progenitors of many coronaviruses. Targeted wildlife surveillance is needed to identify the factors involved in viral perpetuation within natural host populations, and drivers of interspecies transmission. We monitored a natural colony of Egyptian rousette bats at monthly intervals across two years to identify circulating coronaviruses, and to investigate shedding dynamics and viral maintenance within the colony. Three distinct lineages were detected, with different seasonal temporal excretion dynamics. For two lineages, the highest periods of coronavirus shedding were at the start of the year, when large numbers of bats were found in the colony. Highest peaks for a third lineage were observed towards the middle of the year. Among individual bat-level factors (age, sex, reproductive status, and forearm mass index), only reproductive status showed significant effects on excretion probability, with reproductive adults having lower rates of detection, though factors were highly interdependent. Analysis of recaptured bats suggests that viral clearance may occur within one month. These findings may be implemented in the development of risk reduction strategies for potential zoonotic coronavirus transmission.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

