Mass-Suite: a novel open-source python package for high-resolution mass spectrometry data analysis
- PMID: 37741995
- PMCID: PMC10517472
- DOI: 10.1186/s13321-023-00741-9
Mass-Suite: a novel open-source python package for high-resolution mass spectrometry data analysis
Abstract
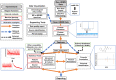
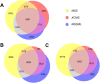
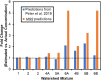
Mass-Suite (MSS) is a Python-based, open-source software package designed to analyze high-resolution mass spectrometry (HRMS)-based non-targeted analysis (NTA) data, particularly for water quality assessment and other environmental applications. MSS provides flexible, user-defined workflows for HRMS data processing and analysis, including both basic functions (e.g., feature extraction, data reduction, feature annotation, data visualization, and statistical analyses) and advanced exploratory data mining and predictive modeling capabilities that are not provided by currently available open-source software (e.g., unsupervised clustering analyses, a machine learning-based source tracking and apportionment tool). As a key advance, most core MSS functions are supported by machine learning algorithms (e.g., clustering algorithms and predictive modeling algorithms) to facilitate function accuracy and/or efficiency. MSS reliability was validated with mixed chemical standards of known composition, with 99.5% feature extraction accuracy and ~ 52% overlap of extracted features relative to other open-source software tools. Example user cases of laboratory data evaluation are provided to illustrate MSS functionalities and demonstrate reliability. MSS expands available HRMS data analysis workflows for water quality evaluation and environmental forensics, and is readily integrated with existing capabilities. As an open-source package, we anticipate further development of improved data analysis capabilities in collaboration with interested users.
Keywords: Mass spectrometry; Non-targeted analysis; Python; Source apportionment; Source tracking; Unsupervised machine learning.
© 2023. The Author(s).
Conflict of interest statement
There are no financial or non-financial competing interests.
Figures
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

